Image
Figure Caption
Fig. s4
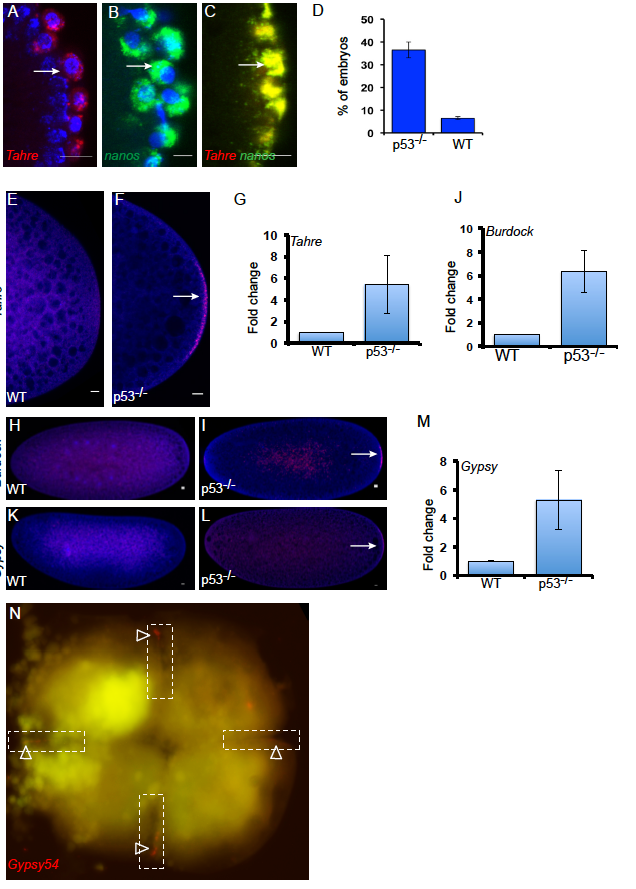
Figure S4. Tahre elements from the oocyte germ plasm persist in the embryonic germline. Related to Figures 3 and 4.
Confocal sections at the cortex of p53-/- embryos labeled with Tahre (A), nanos (B), and co-localization of Tahre with nanos (C), (indicated by arrows) showing that similar to nanos, Tahre transcripts remain highly associated to the pole cells. Embryos 0-3 hr produced from either p53-/- females crossed with WT males or from WT females mated to p53-/- males were scored for the presence of Tahre FISH signal at the pole plasm (D). The maternal effect seen in these reciprocal crosses verifies that Tahre transcripts present in the presumptive germ cells were maternally derived. Tahre, Burdock and Gypsy retroelements are transmitted to the embryonic pole plasm. Confocal images of RNA in situ hybridization of Tahre (E-G), Burdock (H-J) and Gypsy (K-M) on WT and p53-/- (0-2 h) embryos. FISH signal at the pole plasm is shown by arrow. Gypsy transcripts were also detected in the pole plasm of early embryos and could occur below detection limits in the oocyte germ plasm [S5, S6]. Note FISH signals are at the pole plasm in p53-/- embryos. Bar graph quantifies the fold change of the FISH positive signal at the posterior pole of p53-/- embryos relative to WT (0-2 h) from three biological replicates, n=50. DAPI counterstain (blue). Gypsy54 mRNAs localize to the cleavage plane of the Zebrafish embryos. Gypsy54 RNA in situ hybridization on the 4 cell staged p53-/- Zebrafish embryos (N). Dotted lines denote the cleavage plane and arrowheads indicate Gypsy54 signal at the presumptive germline. Scale bars: 10 μm. Error bars represent ±SD.
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Curr. Biol.