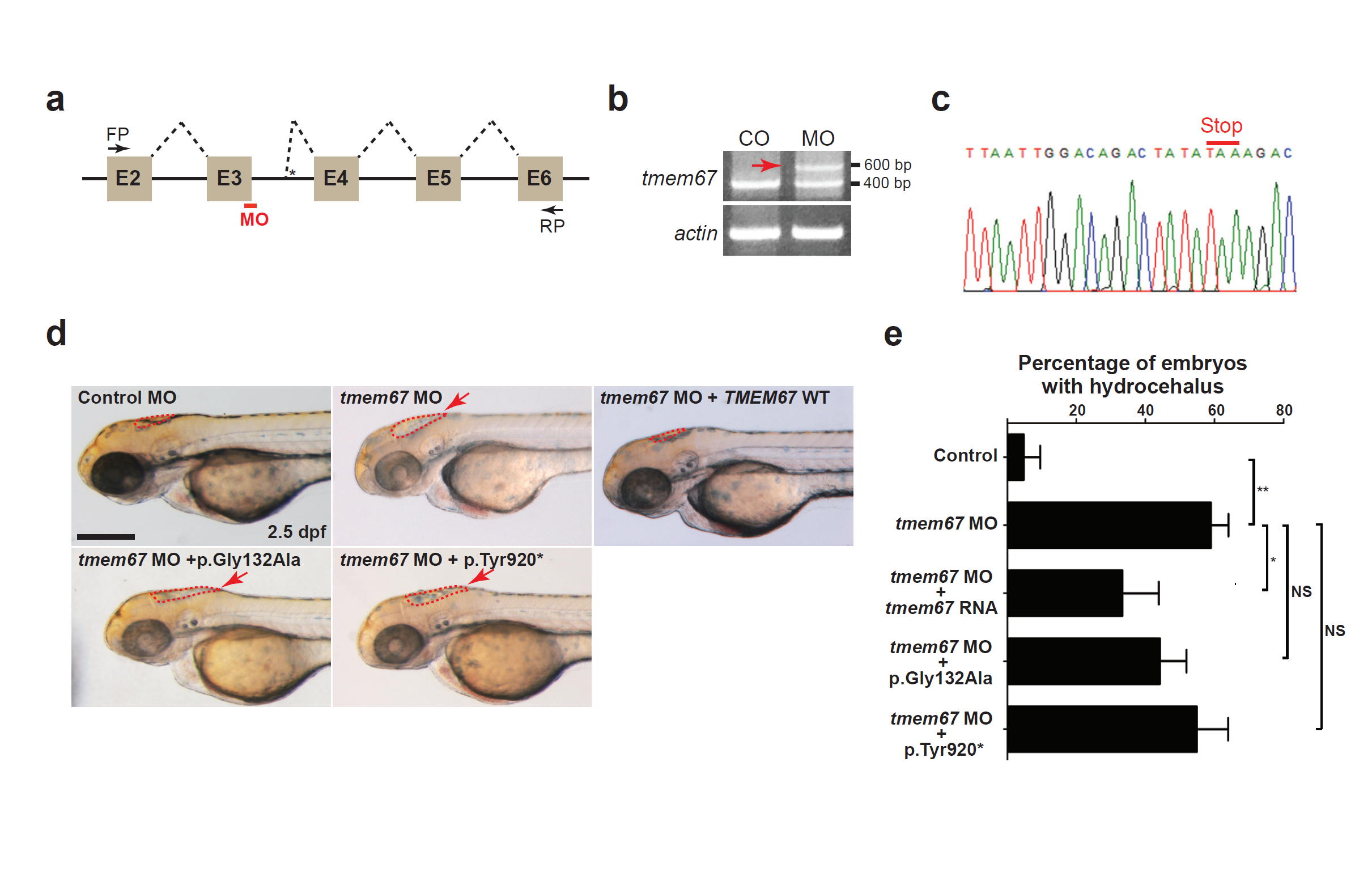
Fig. S2
TMEM67 mutants show decrease in protein function. (a) A schema showing the binding site of tmem67 splice-blocking morpholino (MO [e3i3]). Dashed lines indicate splicing events. Not drawn to scale. E: exon; FP: forward primer; RP: reverse primer. (b) One-cell stage zebrafish embryos were injected with tmem67 MOs and processed for RT-PCR. Αctin was used as a loading control. Arrow indicates a PCR product with deletion of exon3 and retention of a part of intron3. (c) An electropherogram of an exon junction area of the PCR product indicated by arrow in (b). (d) One-cell stage zebrafish embryos were injected with either control MOs or tmem67 MOs [e3ie] and imaged at 2.5 days post-fertilization (dpf). For a rescue experiment, one-cell stage zebrafish embryos were sequentially injected with tmem67 MO [e3i3] and RNA encoding the indicated TMEM67 mutant and imaged at 2.5 dpf. Dashed lines mark the hindbrain ventricles. Arrows indicate enlarged ventricles (hydrocephalus). p.Tyr920* represents p.Tyr920ThrfsX40. Scale bar = 100 μm. (e) Embryos with hydrocephalus were counted in each group in (d). The hindbrain ventricles larger than 5,000 μm2 were considered to be hydrocephalic. *: P < 0.05; ***: P < 0.001 by the two-tailed Student’s t-test; NS: not significant. Number of larvae used in the analysis of each group is over 40.