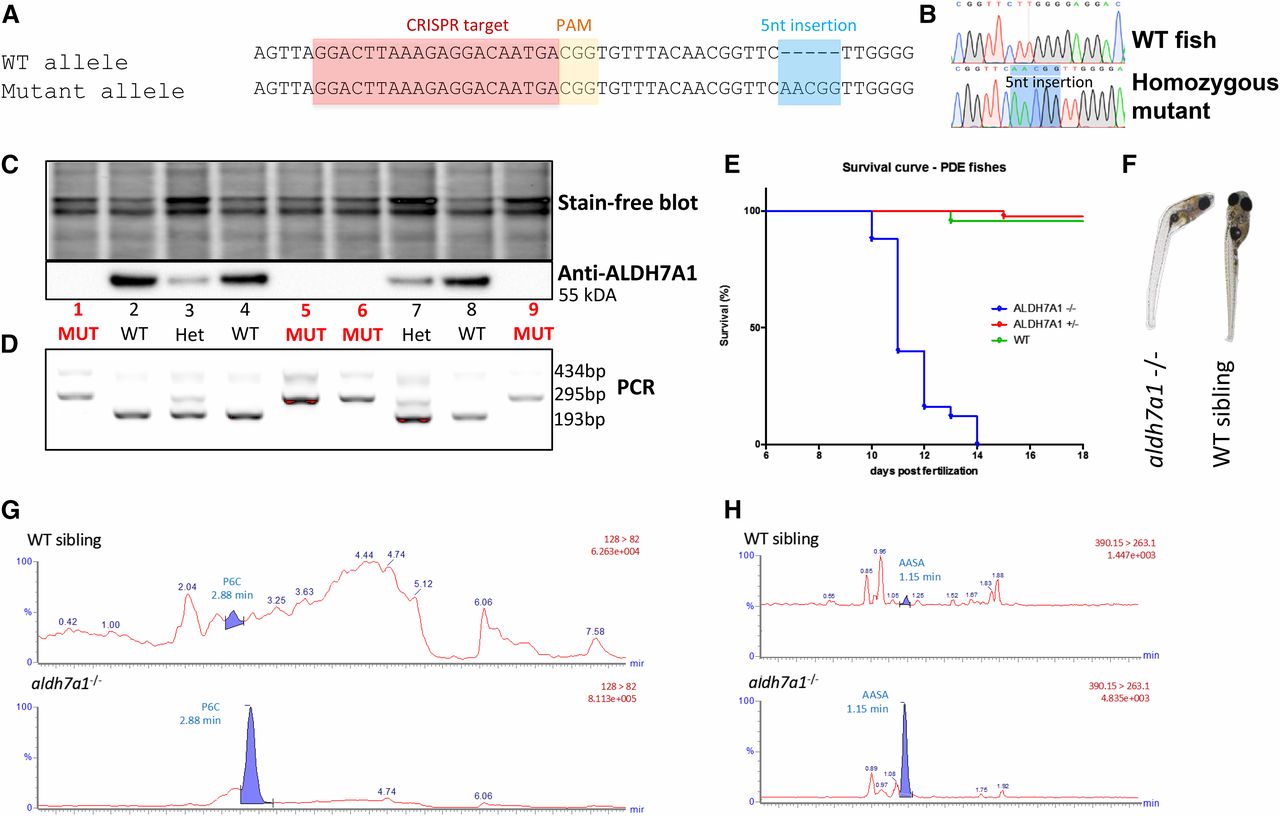
Fig. 2 Development of an aldh7a1−/− zebrafish model by CRISPR/Cas9. The allele aldh7a1ot100 contains a 5-nt insertion mutation (A), which leads to a premature stop codon and an N-terminal-truncated Aldh7a1 protein product. Sequencing chromatograms from F3 larvae show the WT and homozygous mutant (B) patterns. Western blot detected the Aldh7a1 protein in WTs, with ∼50% reduction and complete loss of expression in heterozygous and in the homozygous mutants, respectively (C). PCR-based genotyping (D) using DNA extracted from the fins of the larvae shown in (C) were consistent with the western findings. Kaplan–Meier survival plot showing early death in the homozygous mutants (11–14 dpf), n = 12 aldh7a1−/−, n = 18 aldh7a1+/−, n = 10 aldh7a1+/+ (E). Curved body phenotype observed at 11–14 dpf in the mutants after seizure onset (F). Representative liquid chromatography-mass spectrometry analysis of 7 dpf single larva (n = 6 per genotype); polar metabolite extracts revealed detection and accumulation of the PDE biomarkers P6C and AASA exclusively in the mutants (G and H bottom). No P6C or AASA signal was obtained in the extracts from WT or heterozygous siblings (G and H top). AASA, aminoadipate semialdehyde; CRISPR, clustered regularly interspaced short palindromic repeat; dpf, days postfertilization; HET, heterozygote; MUT, mutant; P6C, piperideine 6-carboxylate; PAM, protospacer adjacent motif; PDE, pyridoxine-dependent epilepsy; WT, wild-type.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Genetics