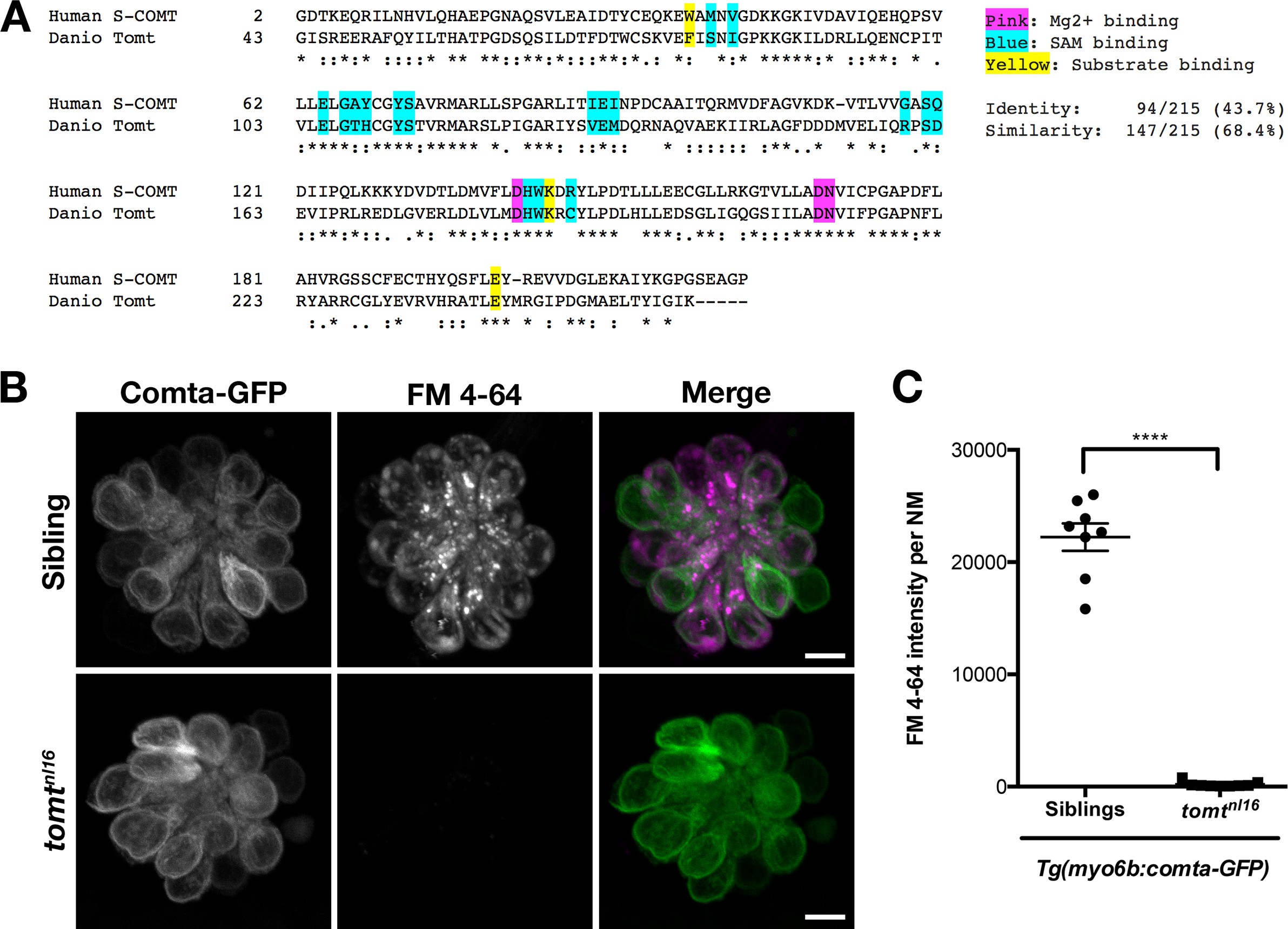
Fig. 7
Comta-GFP cannot restore basal MET channel activity in mercury mutant hair cells.
(A) Alignment between the putative enzymatic domains of human S-COMT (amino acids 2–221, Accession # NP_009294) and zebrafish Tomt (amino acids 43–259, Accession # ANO40802). Residues involved in Mg2+-binding (pink), S-adenosylmethionine (SAM) binding (blue), and catechol substrate binding (yellow) are indicated, as determined by the crystal structure for human S-COMT (PDB_3BWM). Alignment legend - star (*) = conserved; colon (:) = conservative change; period (.) = semi-conservative change. (B) Representative images of Comta-GFP (left panels), FM 4–64 (middle panels) and merged GFP and FM 4–64 channels (right panels) in lateral line NMs of Tg(myo6b:comta-GFP) wild-type siblings and tomtnl16 mutants at 6 dpf. (C) Quantification of FM 4–64 fluorescence intensity per NM for 6 dpf Tg(myo6b:comta-GFP) siblings (n = 5 larvae, 8 NMs) and tomtnl16 mutants (n = 3 larvae, 9 NMs). Error bars are the mean ± SD. Asterisks indicate p<0.0001 by unpaired, two-tailed t-test. Scale bars = 5 µm in B, C.