Fig. S1
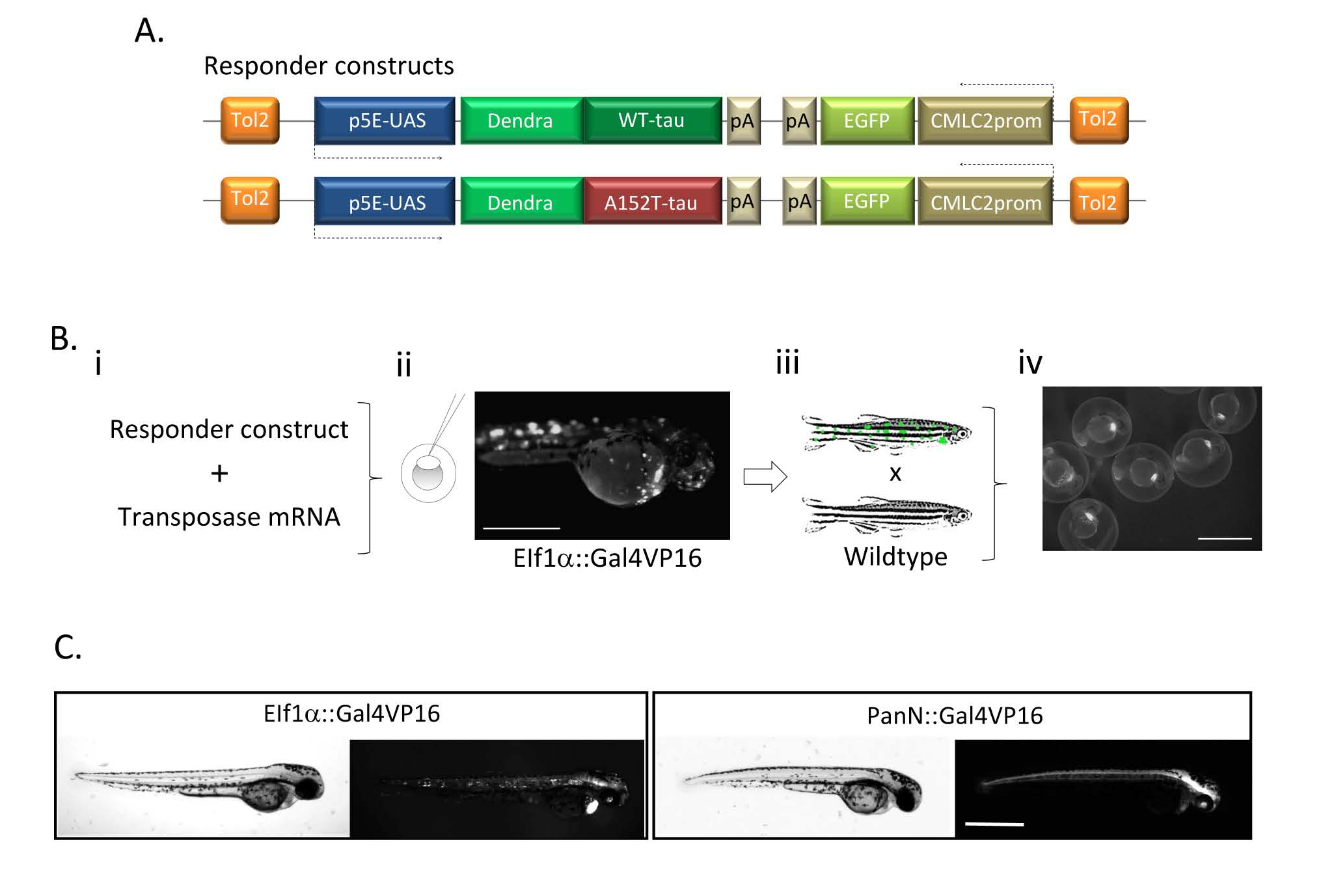
Generation of transgenic Dendra-tau zebrafish: (A) The responder construct comprises the gene encoding human wildtype or mutant A152T tau fused to the sequence encoding the photoactivatable protein Dendra, downstream of UAS. The transgene also contains EGFP driven by the cardiac myosin light chain (CMLC2) promoter in the reverse orientation. The Gal4 driver constructs contain the ubiquitous (EIF1α) or panneuronal (PanN) zebrafish promoters driving the expression of Gal4-VP16, which binds to the UAS on the responder construct. (B) To generate transgenic fish, the responder construct was injected together with the Tol2 mRNA (Bi) to facilitate random integration of the construct into the zebrafish genome resulting in mosaic founder embryos (Bii, scale bar represents 500 μm). Mosaic Dendra-tau positive larvae were identified at 3 d.p.f, raised and outcrossed with wild-type fish (Biii). Transgenic offspring were identified by green hearts i.e. expression from the CMLC2::EGFP reporter (Biv). Scale bar represents 1mm. (C) Dendra-tau expression patterns in the offspring of UAS::Dendra-tau fish crossed with EIF1::Gal4VP16 and PanN:Gal4VP16 driver fish at 48 hours post-fertilisation (h.p.f). Dendra-tau driven by EIF1::Gal4VP16 results in a ubiquitous but mosaic expression of Dendra-tau. Such crosses were used for clearance assays in Fig. 5A,B,G&H, Fig. 6A&B and Fig 7A&B. Ubiquitous transgene expression in offspring from crosses to beta-actin::Gal4VP16 driver fish were used for proteasome assays in Fig. 6F&G (Dendra-tau driven by beta-actin::Gal4VP16 not shown). All other experiments use Dendra-tau driven by PanN::Gal4VP16Scale bar represents 500 μm.