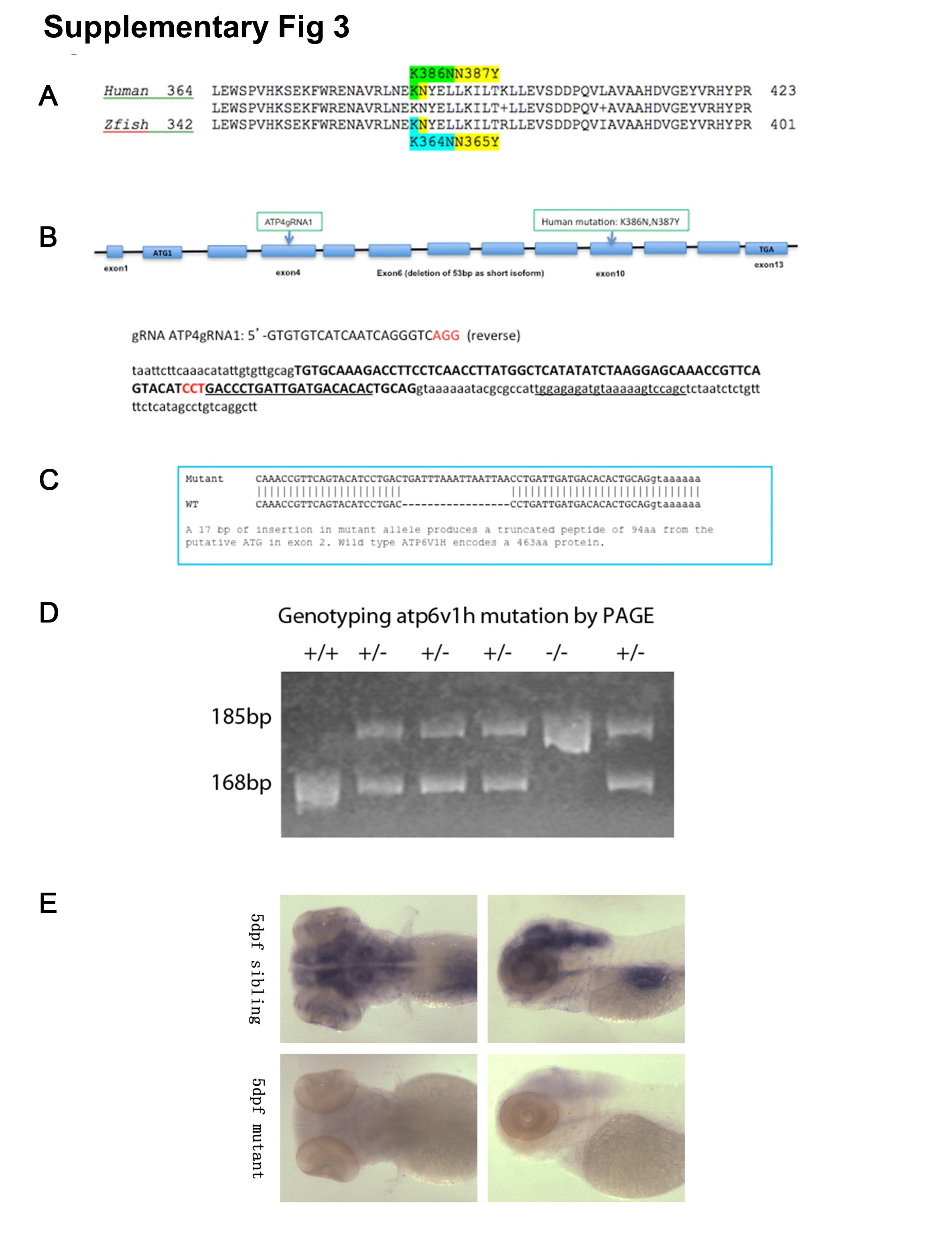
Fig. S3
Generation and Genotyping of atp6v1h zebrafish mutants.
Protein alignment of ATP6V1H in human (NP_057025.2) and zebrafish (NP_775377.1) shows high homology (~85%). More importantly, the region where the mutation is located is highly conserved (A). Using CRISPR/Cas9, guide RNA (gRNA) targeting exon 4 of zebrafish atp6v1h was co-injected with Cas9 mRNA into zebrafish embryos and detected by T7 endonuclease digestion. Sequence of gRNA and target is shown (B). Several founders with indels were screened for germline transmission, and the one with 17bp insertion was used throughout the study; The 17bp insertion is predicted to produce a termination codon 94 amino acids after initiation of start codon (C). Genotyping done by polyacrylamide gel electrophoresis (PAGE) analysis demonstrating a 168 bp band for wild type allele, and 185 bp band for mutant allele; representative results for wild type (+/+), heterozygous (+/-), and homozygous (-/-) embryos are shown (D). Using probes specific for atp6v1h, RNA in situ hybridization analysis of atp6v1h showed expression in the head region in wild type embryos, while the expression is nearly absent in homozygous atp6v1h embryos, suggesting that the 17bp insertion may lead to nonsense-mediated decay (NMD) of mRNA and result in loss-of-function of this gene (E).