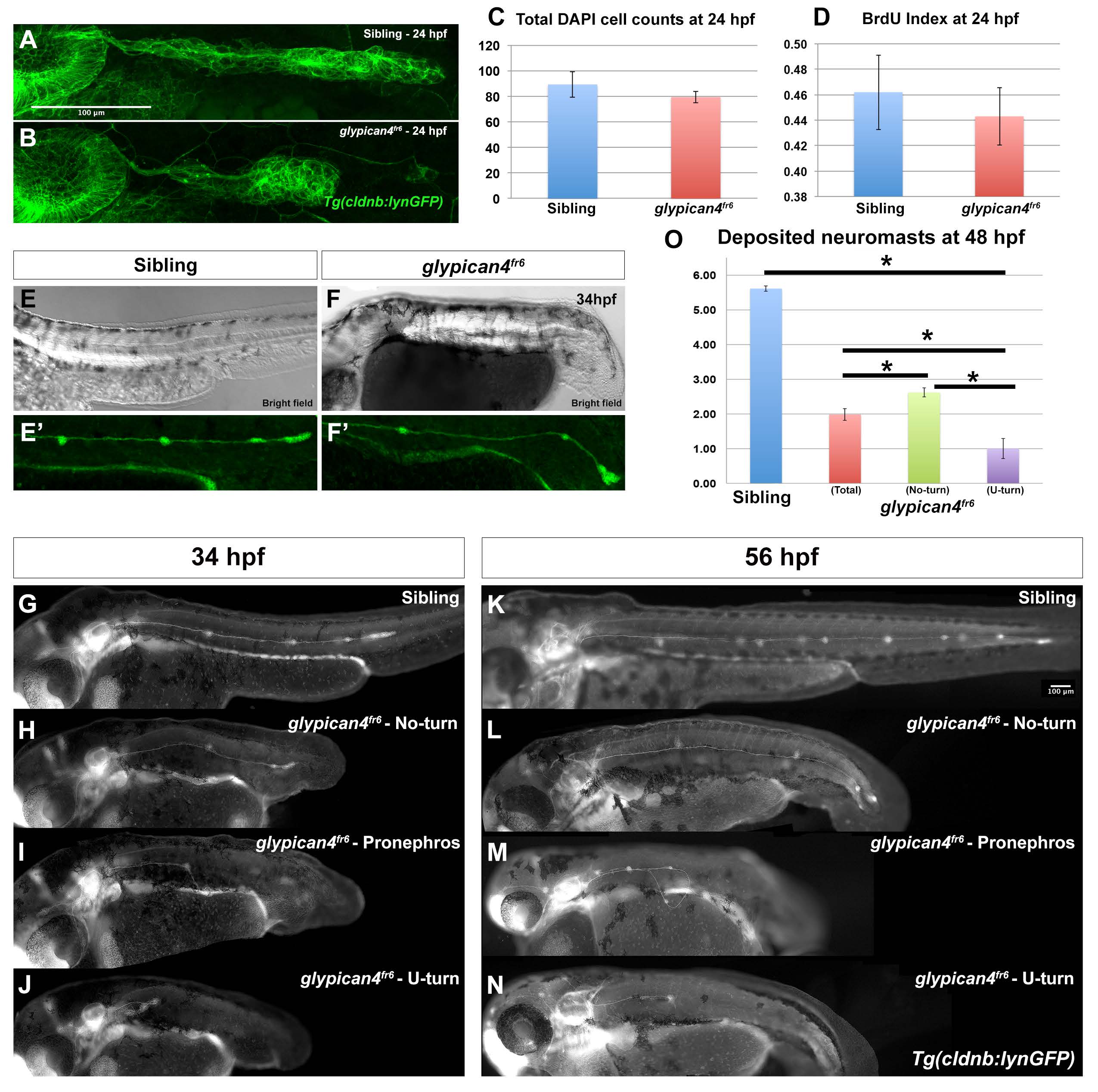
Fig. S1
glypican4fr6 primordia have morphology defects and deposit fewer neuromasts. (A-B) Compared to control siblings where the primordium has an elongated ovoid shape (A), glypican4fr6 mutant primordia are more rounded and of smaller size (B). (C) Cell counts by DAPI staining show no significant differences between sibling and glypican4fr6 mutant primordia at 24 hpf (p-value (t-test)=0.327). (D) The reduced size of the 24 hpf glypican4fr6 mutant primordia is not due to reduced proliferation (p-value (t-test)=0.682). (E-F) Bright field images of a sibling control and a glypican4fr6 mutant embryo at 34 hpf depicting the embryo trunk phenotypes. (E′-F′) Same embryos represented in (E) and (F) under fluorescent light showing the lateral line primordium migrating along the trunk. No-turning glypican4fr6 mutant primordia reach the tail tip shortly after 34 hpf, whereas the sibling primordium has just passed the yolk extension. (G-J) Lateral views of 34 hpf embryos depicting representative phenotypes of sibling (G) and the three phenotypes of glypican4fr6 mutant primordia migration (H-J). (K-N) Lateral views of 56 hpf embryos depicting representative phenotypes of sibling (K) and the three phenotypes of glypican4fr6 mutant primordia migration (l-N). (O) Quantification of deposited neuromasts at 58 hpf in sibling versus glypican4fr6 mutants (siblings=5.61±0.08 vs. glypican4fr6(total)=1.98±0.17 neuromasts, n=56 embryos. sibling vs. glypican4fr6 (total) p-value=1.7×10−36; sibling vs. glypican4fr6 (No-turn) p-value=2.38×10−35; sibling vs. glypican4fr6 (U-turn) p-value=1.77×10−33). Error bars in (C), (D) and (O) represent standard errors and (*) represent statistically significantly phenotypes.
Reprinted from Developmental Biology, 419(2), Venero Galanternik M., Lush, M.E., Piotrowski, T., Glypican4 Modulates Lateral Line Collective Cell Migration Non Cell-Autonomously, 321-335, Copyright (2016) with permission from Elsevier. Full text @ Dev. Biol.