Fig. 1
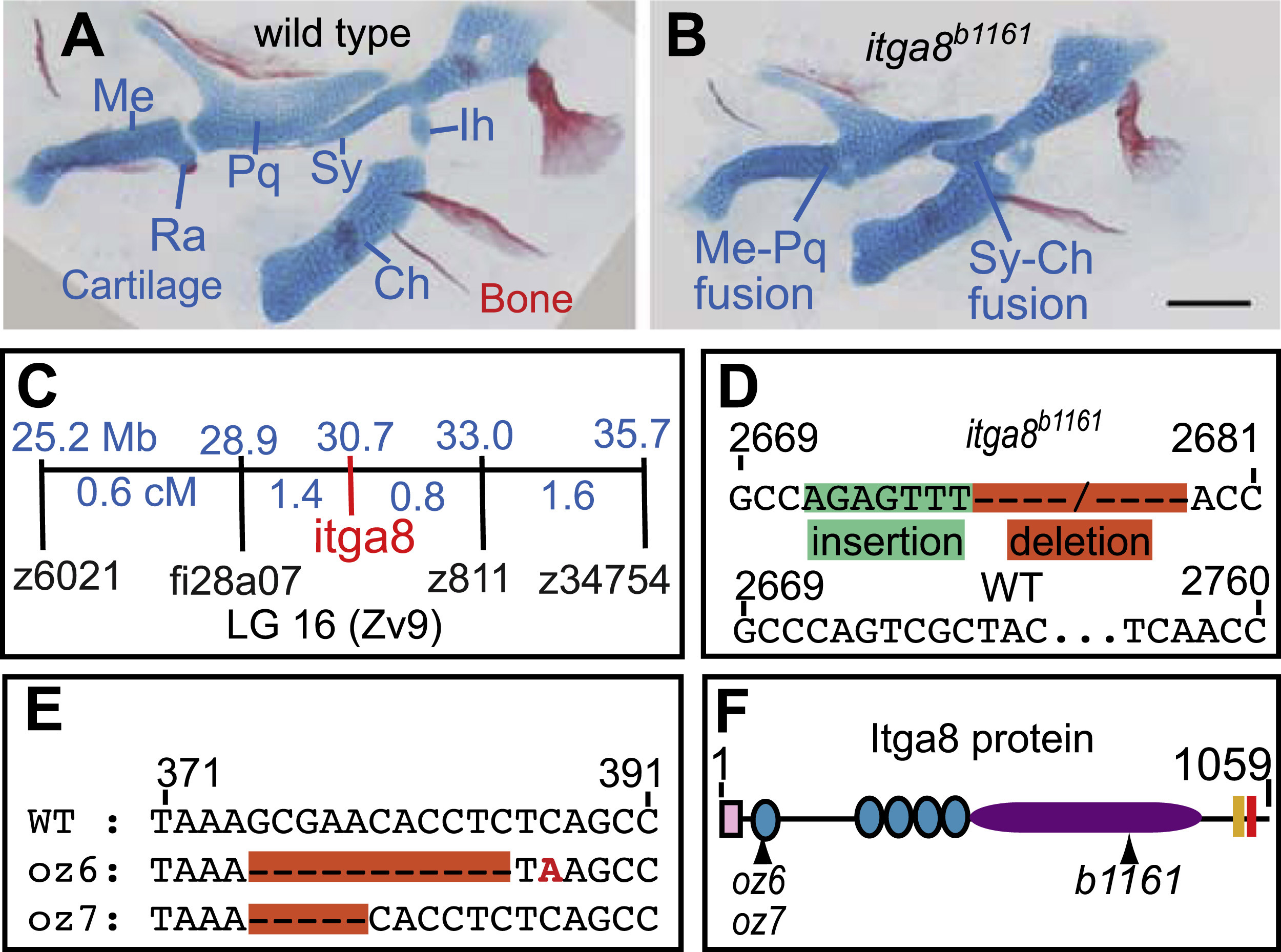
Skeletal defects in b1161 mutants are caused by lesions in itga8. (A, B) Alcian and alizarin staining of (A) wild type and (B) itga8b1161 homozygotes shows skeletal defects caused by itga8 mutations. Homozygous itga8 mutants often show cartilage fusions in the first two pharyngeal arches and defects in symplectic length. (C) Linkage analysis reveals no recombinants (0/696 individuals) between b1161 and the itga8b1161, placing them at the same map position. Map distances (blue) of additional markers from b1161 are shown above in Mb and below in cM. (D) Sequence of the itga8b1161 lesion reveals a 7 bp insertion in exon 25 (green), followed by a 79 bp deletion (orange) (wild type: GenBank JN399198, b1161: GenBank JN399198). This lesion results in protein truncation after amino acid 845 of 1059 with the predicted addition of 21 aberrant amino acids (EFTHWSWRPRLFRTLQSYWAS). (E) Sequence of two CRISPR-induced itga8 lesions, itga8oz6 (11 bp deletion) and itga8oz7 (5 bp deletion), reveal that both introduce frameshifts after amino acid 79 of 1059; itga8oz6 causes an immediate stop codon after the frameshift, while itga8oz7 introduces 30 aberrant amino acids (HLSAGDCGGRSGVLLPLAGIRPRLLPPDPL) before terminating. (F) Itga8 protein diagram with locations of oz6, oz7, and b1161 mutations along with predicted protein motifs. Protein motifs are designated as follows: signal sequence (pink box), integrin beta domains (teal circles); integrin alpha domain (purple oval), transmembrane domain (peach box), and an intracellular integrin domain (red box). Cartilage abbreviations: Meckel’s (Me), Retroarticular process (Ra), Palatoquadrate (Pq), Symplectic (Sy), Ceratohyal (Ch), Interhyal (Ih). Scale bar (100 µm) in B also applies to A.
Reprinted from Developmental Biology, 416(1), Talbot, J.C., Nichols, J.T., Yan, Y.L., Leonard, I.F., BreMiller, R.A., Amacher, S.L., Postlethwait, J.H., Kimmel, C.B., Pharyngeal morphogenesis requires fras1-itga8- dependent epithelial-mesenchymal interaction, 136-48, Copyright (2016) with permission from Elsevier. Full text @ Dev. Biol.