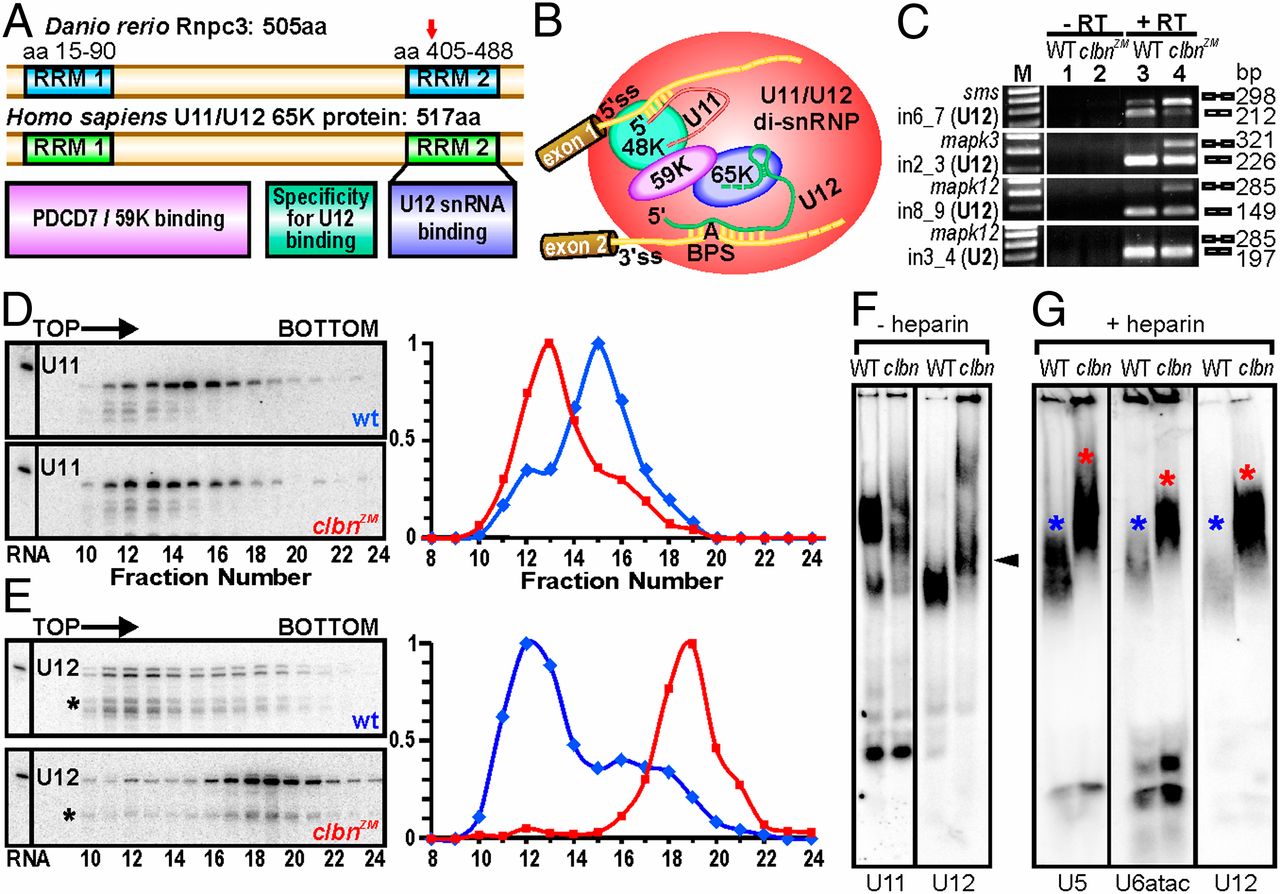
Fig. 3
Zebrafish Rnpc3 functions in the U12-type spliceosome. (A) Domain structure of zebrafish Rnpc3 and its human ortholog RNPC3, or U11/U12 di-snRNP 65-kDa protein (www.uniprot.org, accession no. Q96LT9). Red arrow in the C-terminal RRM denotes the approximate position of the premature stop codons in all clbns846 isoforms. (B) Schematic of the human U11/U12 di-snRNP, with RNPC3 (65 K) bridging U12 snRNA and the 59 K protein (28). BPS, branch point sequence; “A” represents the branch point adenosine. (C) RT-PCR analysis of three different transcripts (sms, mapk3, mapk12) shows retention of U12-type introns (lane 4, Top three panels) in clbns846 (5 dpf), but not of a U2-type intron (Bottom). Lanes 1 and 2 show –RT controls. (D and E) Glycerol gradient/Northern analysis revealing differential sedimentation of U11 (D) and U12 (E) snRNA-containing snRNPs in WT and clbnZM extracts. Direction of sedimentation is left to right. Lane 1 contains total RNA. Full-length U11 or U12 signals were quantified using ImageQuant and expressed as a percentage of the fraction with the highest intensity. U12 snRNA showed persistent specific degradation (asterisks) without affecting its migration profile. (F) Northern analysis of WT and clbns846 extracts resolved on 4% (80:1) native polyacrylamide gels and probed for U11 and U12 snRNAs. The predominant U11 snRNPs are disrupted in clbn compared with WT and the U12-containing particles are heavier and migrate more slowly (black arrowhead). (G) Northern analysis of WT and clbns846 extracts resolved on native gels and probed for U5, U6atac, and U12 snRNAs shows retarded bands in clbn (red asterisks) compared with WT (blue asterisks). The same larval lysate was used in F and G. Data are representative of a total of 10 (D and E) and 5 (F and G) biological replicates.