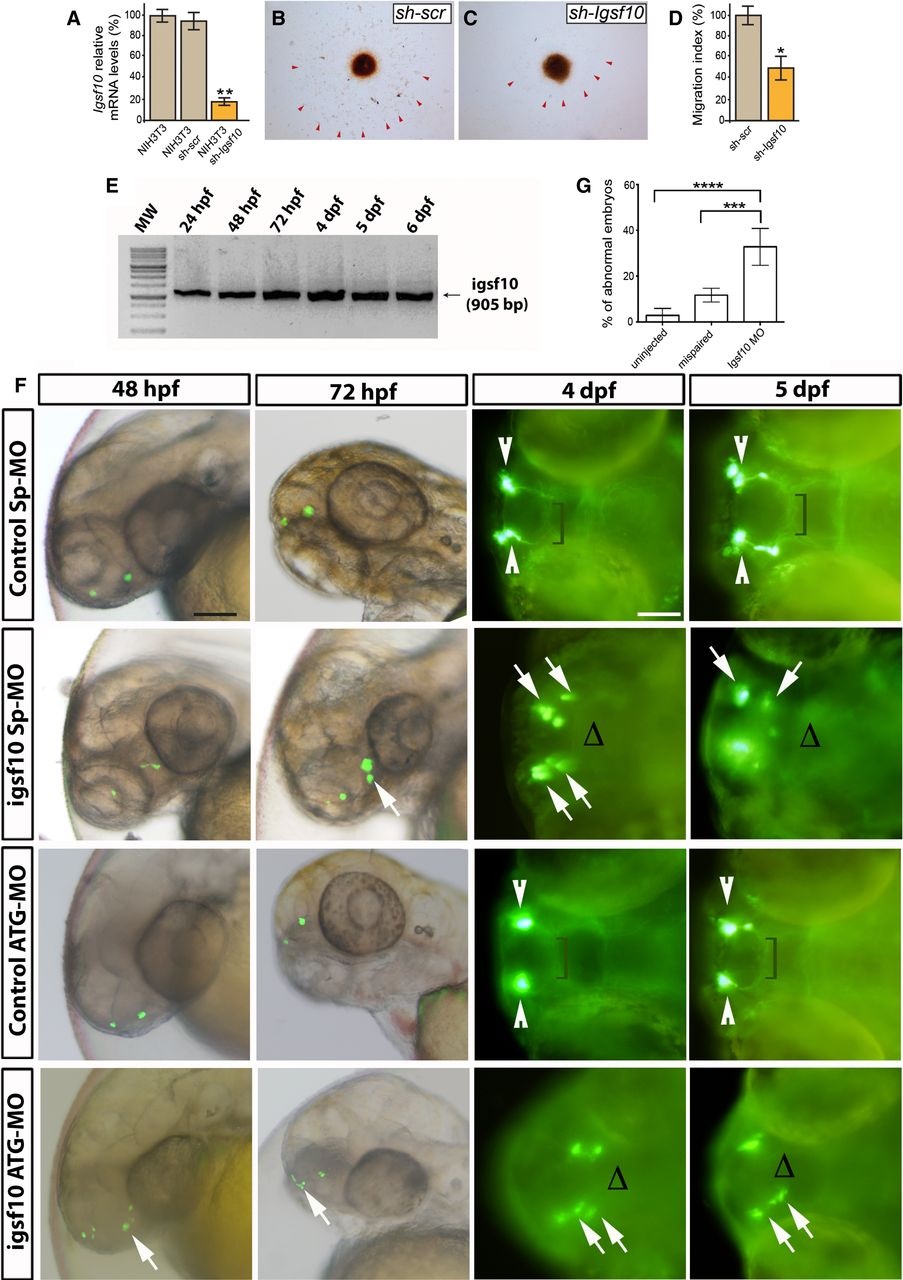
Fig. 5
Effect of Igsf10 knockdown on GnRH neuronal migration
A. Levels of Igsf10 expression in native NIH3T3 cells, and cells stably expressing a scrambled (shscr) or Igsf10 (shIgsf10) shRNA.
B, C Migration of GN11 cells from aggregates into NIH3T3 shscr (B) or shIgsf10 (C).
D. Analysis of the migration index (see Materials and Methods).
E. RT–PCR analysis in zebrafish of total embryos and larvae showing the expression of Igsf10 mRNA at different timepoints. Expected band size: 905 bp.
F. Representative timelapse analysis of mispair control (control SpMO and control ATGMO) and Igsf10 morpholinos (Igsf10 SpMO and Igsf10 ATGMO)injected Tg(gnrh3:EGFP) embryos and larvae, at different timepoints. Images show lateral view of live larvae head (48 and 72 hpf) and dorsal view (4 and 5 dpf) of live larvae head; anterior is left. White arrowheads indicate normal GnRH3 neuron clusters in the olfactory area of control embryos. Brackets show the extension of the projections toward the hypothalamus. White arrows indicate examples of abnormal GnRH3 neurons scattered in the olfactory area of morphants accompanied by the lack of projections toward the hypothalamus, indicated with Δ (hpf, hours postfertilization; dpf, days postfertilization). Scale bar, 100 µm (panels at 48 and 72hpf in F), 150 µm (panels at 4 and 5 dpf in F).
G. Effect of Igsf10 splicesite morpholino injection observed at 48 h. Quantification of the percentage of embryos showing an abnormal GnRH3-neuron phenotype was significantly higher in Igsf10 splicesiteMO (SpMO)injected embryos compared to relative controls.