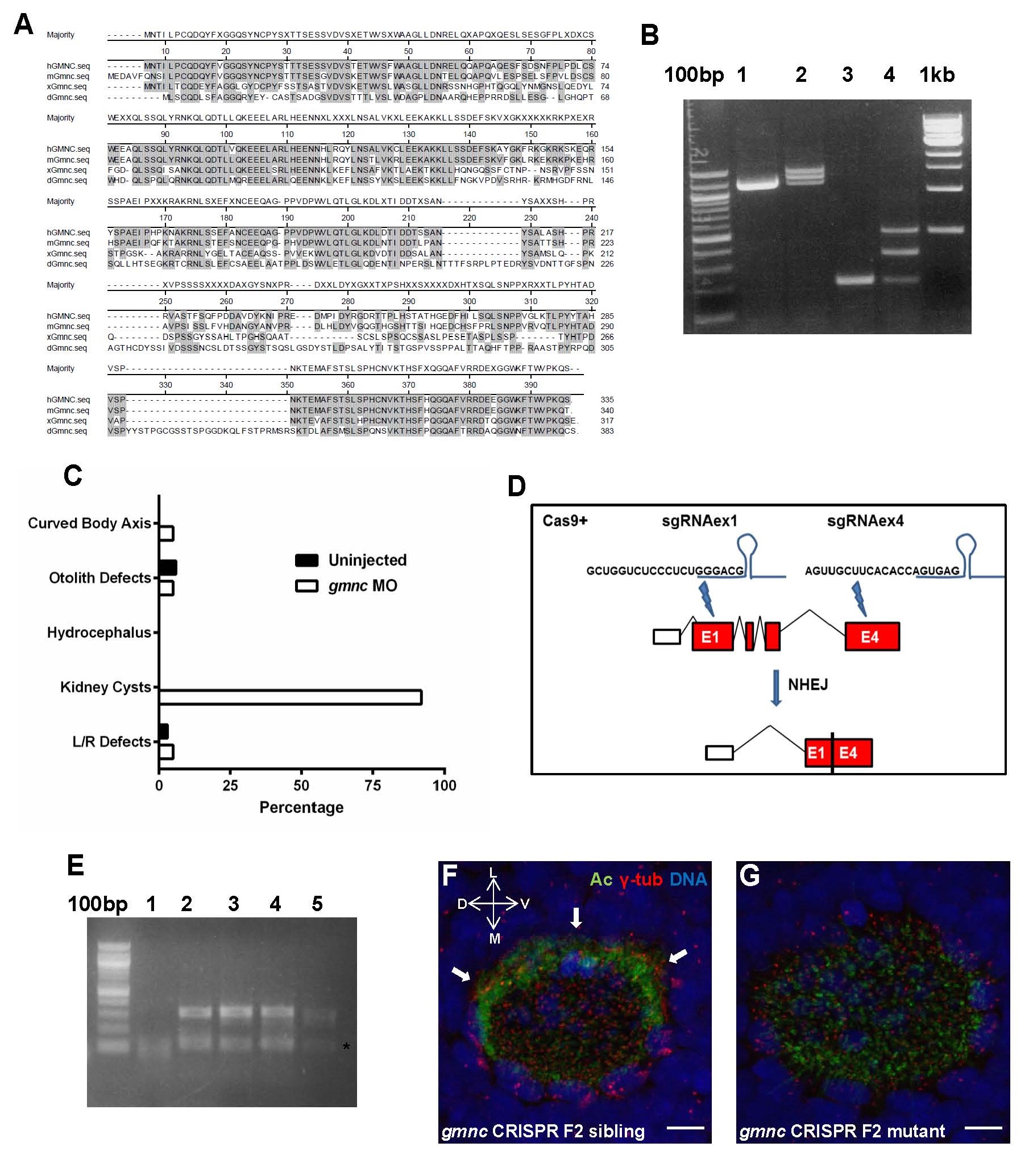
Fig. S1
Sequence comparisons of Gmnc orthologs, efficacy of splicing inhibition by the zebrafish gmnc splice blocking MO, scoring of ciliary dysfunction-associated phenotypes in gmnc MO embryos, CRISPR/Cas9-mediated deletion in the zebrafish gmnc genomic region, and defects in cilia formation in the embryonic nasal placode in gmnc mutants
(A) Gmnc amino acid sequences from human (hGMNC), mouse (mGmnc), Xenopus (xGmnc) and zebrafish (dGmnc) proteins were aligned using the Clustal W method. Regions of homology among the four species are shaded.
(B) The gmnc splice morpholino was designed to inhibit splicing at the exon 2/intron2-3 junction of the zebrafish gmnc pre-mRNA. To verify its efficacy, cDNAs from 24 hpf wild-type (lane 1) and gmnc morpholino injected (lane 2) embryos were used in end-point PCRs to amplify the 1.1kb-long gmnc coding sequence. A single band of the expected size was amplified from the wild-type, while 3 bands of incremental sizes were amplified from the morphant. To resolve the identity of these bands, primers were designed to amplify across the 319 bp-long intron 2-3, with an expected PCR product size of ~200 bp from the wild-type (lane 3). This strategy revealed two additional bands in the morphants (lane 4), with the ~500bp band corresponding to the PCR product that includes the retained intron 2-3.
(C) Five ciliary phenotypes were scored in uninjected zebrafish embryos or gmnc MO-injected embryos: The extent of body axis curvature, otolith defects in the inner ear (other than two), swelling of the brain ventricles (hydrocephalus), kidney cysts, and heart looping directionality (L/R defects) (n=40). The morphants did not exhibit any overt ciliary dysfunctional phenotypes other than the prevalent formation of kidney cysts.
(D and E) Schematic showing the CRIPSR/Cas9-mediated genome modification at the zebrafish gmnc locus designed to excise a region between exons 1 and 4, using two guide RNAs (sgRNAex1 and sgRNAex4). After non-homologous end-joining (NHEJ) repair, the modified genomic locus would only contain part of exon 1 and exon 4 fused, lacking the intervening sequences (D). Primers flanking the deletion region were used to identify fish carrying the deletion (lanes 2 to 5, with an expected PCR product size of ~350bp, which was then sequenced to verify the region of the deletion) versus wild-type animals (lane 1, where the ~350bp product should be absent) (E). The lower bands (asterisk) are probably non-specific PCR product(s).
(F and G) A prominent band of MCCs (indicated by arrows) decorate the lateral circumference of the embryonic nasal placode at 72hpf (F). This MCC band was completely lost in the gmnc mutant embryos (100%, n=10) (G). Acetylated-tubulin (Ac; green), γ- tubulin (red), DAPI (blue). D: dorsal; V: ventral; L: lateral; M: medial. Scale bars: 5µm.