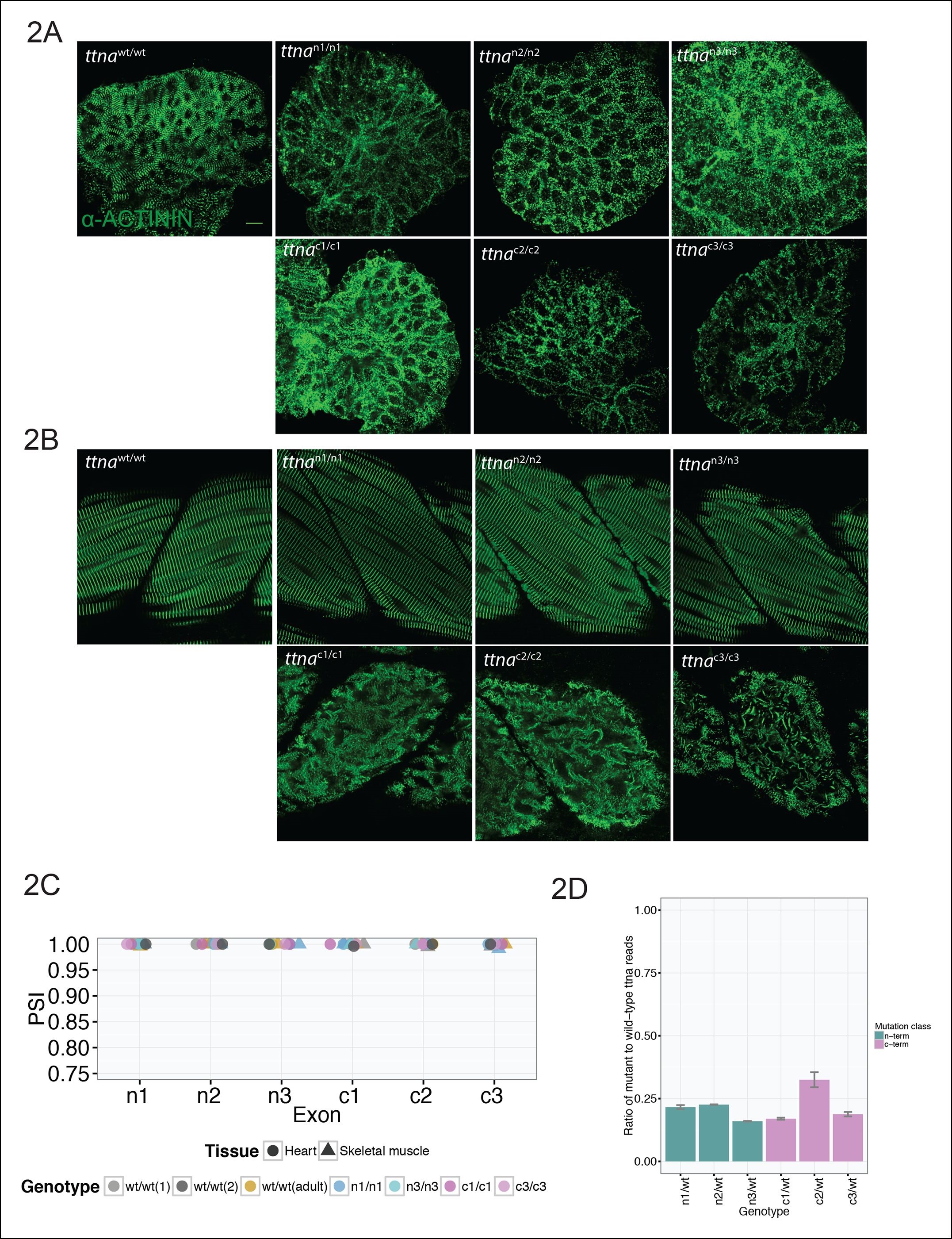
Fig. 2
C-terminal ttna truncations result in a severe skeletal muscle phenotype while N-terminal truncations are indistinguishable from wild-type.
Fixed heart (A) and skeletal muscle (B) samples of 72 hpf ttnawt/wt, ttnan/n and ttnac/c mutant embryos were analyzed by immunostaining for α–actinin, which highlights Z-disc architecture. The cardiac sarcomere was disarrayed in all mutants. However, in skeletal muscle ttnac/c mutants demonstrated severe sarcomeric disarray while ttnan/n mutants retained sarcomeric architecture. Scale bar: 10 uµm. (C) All targeted TTN exons are constitutive (i.e. not alternatively spliced) in both cardiac and skeletal muscle. PSI values computed for each mutated exon using RNA-Seq data for dissected hearts and trunk skeletal muscle for various mutant genotypes at 72 hpf. Wild-type fish were analyzed at both 72 hpf and adulthood. Analysis was limited to samples with a sufficient number of exon-exon junction reads to accurately estimate PSI (Pervouchine et al., 2013). (D) Nonsense-mediated decay reduces mutant transcript levels to ~20–25% of wild-type, but does not vary substantially across mutations. Targeted RNA-Seq was used to determine the ratio of reads derived from the mutant allele vs. the wild-type allele in ttnan/wtand ttnac/wt heterozygote mutants, which serves as an estimate of nonsense-mediated decay efficiency.