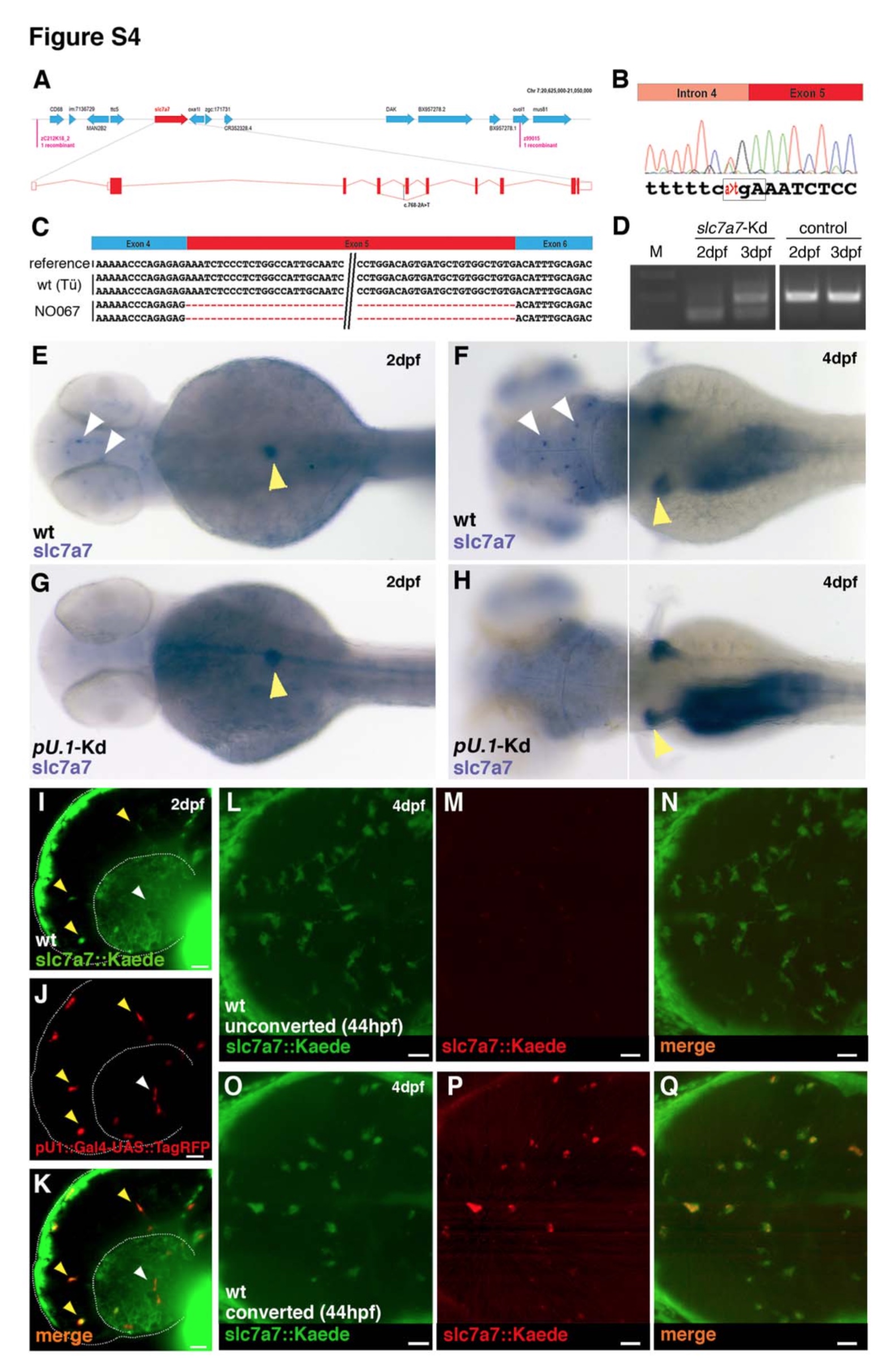
Fig. S4 Slc7a7 is expressed in anteriorly located macrophages and is necessary for the establishment of the microglial population. Related to Figure 3 and Figure 4.
(A) Genetic map of the NO067/slc7a7 locus. (B and C) Sequence chromatogram showing the mutation in the slc7a7 coding sequence (ENSDARG00000055226). Sequence analysis identifies an A to T mutation in the fourth splice acceptor of slc7a7 (B) that leads to skipping of the fifth exon (C). Box in (B) indicates the premature stop codon. (D) RT-PCR analysis using primers flanking the exon5 of the slc7a7 cDNA to confirm the efficacy of the morpholino-mediated knockdown. A band shift is present both at 2 and 3 dpf in injected embryos. (E-H) WISH for slc7a7 in a wildtype (E-F) and a pU.1 morphant (G-H) embryo at 2 dpf and 4 dpf (dorsal view). Knockdown of pU.1 leads to lack of slc7a7+ cells in the head (white arrowheads) but not in pancreas and kidney (yellow arrowheads). (I-K) Lateral view of a representative slc7a7::Kaede;pU.1::TagRFP embryo at 2 dpf. Slc7a7+ cells are in green (I, slc7a7::Kaede) and leukocytes are in red (J, pU.1::TagRFP), (K) overlay. Few cells are both pU.1+ and slc7a7+ (yellow arrowheads). White arrowheads indicate pU.1+ cells only. (L-Q) Dorsal view of a representative brain of a 4 dpf slc7a7::Kaede transgenic embryo not photoconverted (L-N) and photoconverted at 44 hpf (O-Q). (L and O) Kaede green, (M and P) Kaede red, (N and Q) overlay. Scale bars for all images: 25 μm.