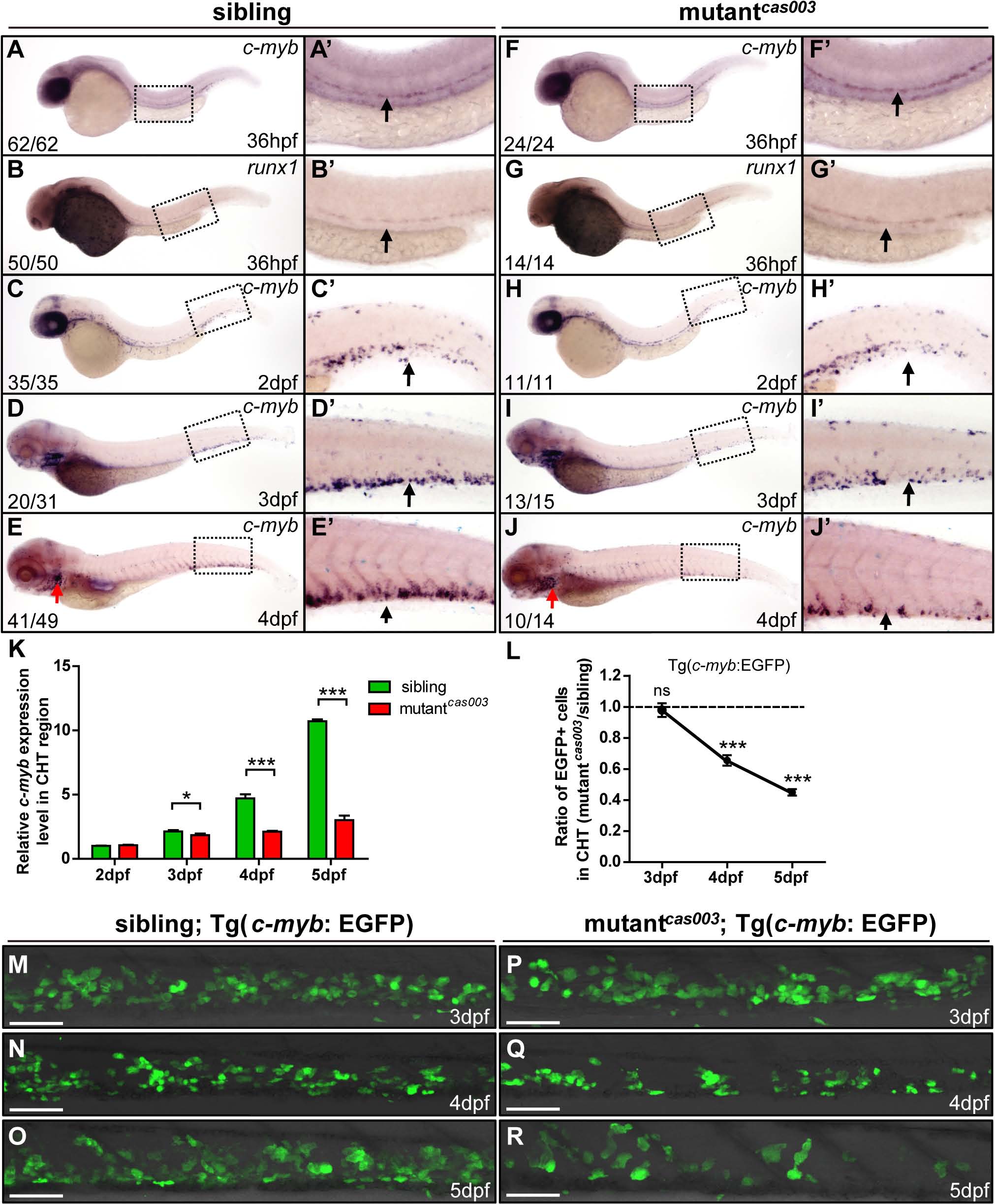
Fig. 2 The number of HSPCs is reduced in the mutantcas003 CHT.
(A-J′) The time-course analysis of definitive HSPCs from 36hpf to 4dpf by WISH with c-myb (A, A′, C-E′, F, F′, H-J′) or runx1 (B, B′, G, G′) indicating the HSPCs are decreased from 3dpf in mutantcas003 embryos comparing to siblings. The penetrance of the indicated phenotype is shown in the bottom left of each panel. (A′-J′) Enlarged views of AGM or CHT regions indicated by the dotted rectangles in the left columns. Black arrows represent the AGM (A′, B′, F′, G′) or CHT (C′-E′, H′-J′); red arrows indicate the thymus. (K) Quantitative PCR analysis of c-myb expression level in the tail region from 2dpf to 5dpf showing the attenuated c-myb expression after 3dpf in mutantcas003 embryos comparing to siblings. Error bars represent SEM. *, p<0.05; ***, p<0.001. (L-R) Immunofluorescence stain of EGFP in the CHT region in mutantcas003; Tg(c-myb: EGFP) embryos (P-R) and siblings (M-O) from 3dpf to 5dpf. The statistics result of the ratio of EGFP+ cell number in the CHT region of mutantcas003 embryos to that in the siblings is indicated in L (n>6 embryos are counted in each panel). These results indicate that EGFP+ HSPCs begin to be decreased obviously in CHT region in mutantcas003 embryos comparing to siblings from 4dpf. Scale bars represent 50µm. Error bars represent SEM; ns, no significance; *** represents p<0.001.