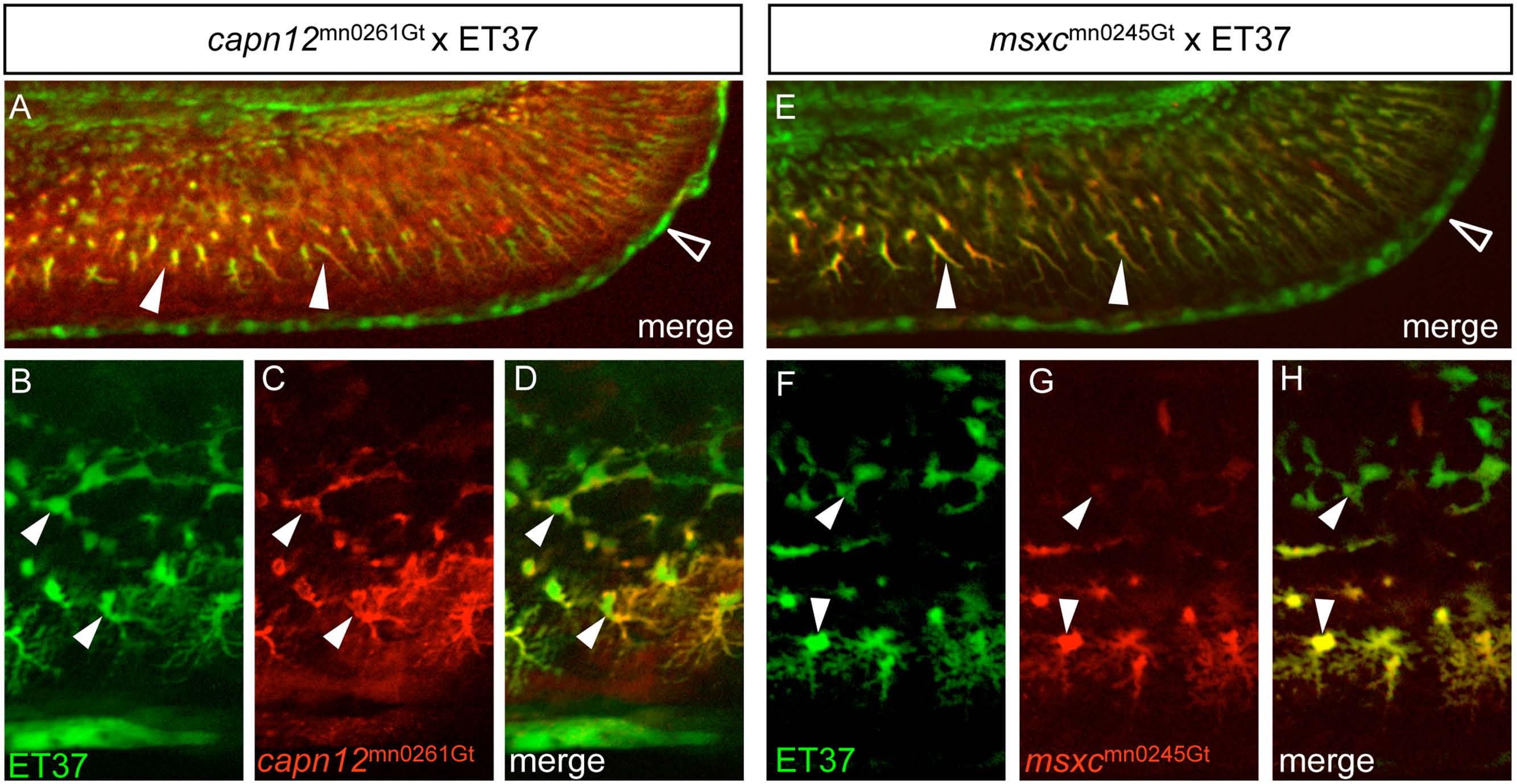
Fig. 3 capn12 and msxc transgenes are co-expressed with a fin mesenchymal cell marker.
(A, E) When capn12mn0261Gt and msxcmn0245Gt are crossed into an ET37 background, both Capn12mn0261Gt-mRFP and MsxCmn0245Gt-mRFP co-localize with ET37 EGFP-positive fin mesenchymal cells (FMCs) (arrowheads). ET37 also labels the cleft cells along the MFF edge (open arrowheads). Neither Capn12mn0261Gt-mRFP nor MsxCmn0245Gt-mRFP localizes to the cleft cells. (B-H) ET37 EGFP labels all FMCs within the fin (arrowheads), both proximal (upper arrowheads) and distal (lower arrowheads), and is present throughout FMC cell bodies and in the extended “arbors” of distal FMCs (lower arrowheads). Both Capn12mn0261Gt-mRFP and MsxCmn0245Gt-mRFP localization show similarities with and differences from ET37 EGFP. (B-D) Both ET37 EGFP and Capn12mn0261Gt-mRFP are expressed in proximal and distal FMCs (arrowheads) but unlike ET37 EGFP, Capn12mn0261Gt-mRFP localizes to the periphery of FMCs (arrowheads). (F-H) MsxCmn0245Gt-mRFP co-localizes with ET37 EGFP, but differs from Capn12mn0261Gt-mRFP. MsxCmn0245Gt-mRFP is much weaker in proximal FMCs (upper arrowheads) than in distal FMCs (lower arrowheads). Unlike Capn12mn0261Gt-mRFP, MsxCmn0245Gt-mRFP subcellular localization is not noticeably distinct from that of ET37 EGFP.