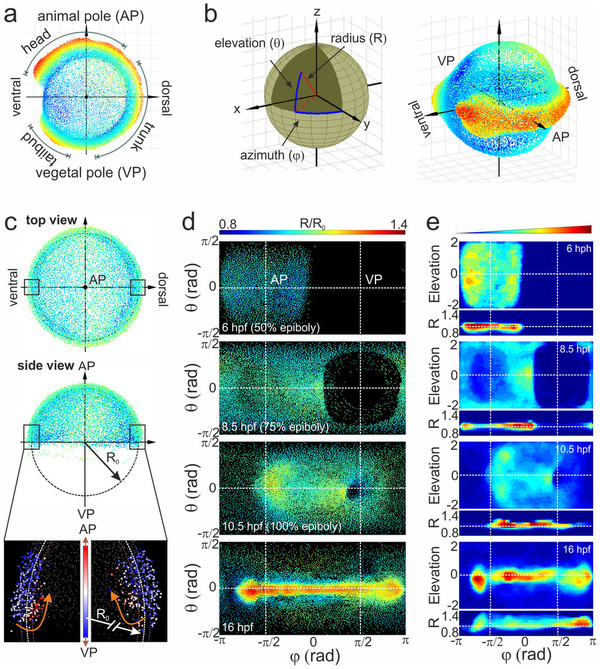
Fig. 2
Alignment of cell coordinates according to the standard embryo position.
(a) Standard embryo representation on the yolk sphere at later developmental time points (16hpf); the color code denotes the distance from the origin. (b) Transformation from Cartesian to spherical coordinates. For the cylindrical projection, the anterior-posterior embryo axis was placed at zero elevation (equator); by setting the dorsal embryo side at zero azimuths, the animal (AP) and vegetal (VP) poles appeared at the π/2 and π/2 azimuthal angles, respectively. (c) Embryo alignment at the early gastrula stage. Top and side views show the symmetrically distribution of cells around the AP. The color code denotes the distance from the origin as in (a). The radius of the embryo sphere R0 is determined according to the border between hypoblast and epiblast cell layers (lower panel of the side view; the color code denotes the movement direction) which display opposite movement at 7hpf towards the AP (red) and VP (blue), respectively. (d) Spherical projection of cell nuclei coordinates from a single embryo on a 2D map using azimuthal and elevation angles. Typical 2D maps of cell nuclei coordinates are plotted at 6, 8.5, 10.5 and 16hpf using the equidistant cylindrical projection. The radial coordinate is color-coded in the range between 0.8 and 1.4. (e) 2D maps of integrated cell nuclei density from a single sample using the area-preserving Gall-Peters (2 sinθ versus Φ) and R versus Φ projection of the same datasets as in (d). The blue-to-red color code denotes the increase of the normalized cell density in arbitrary units.