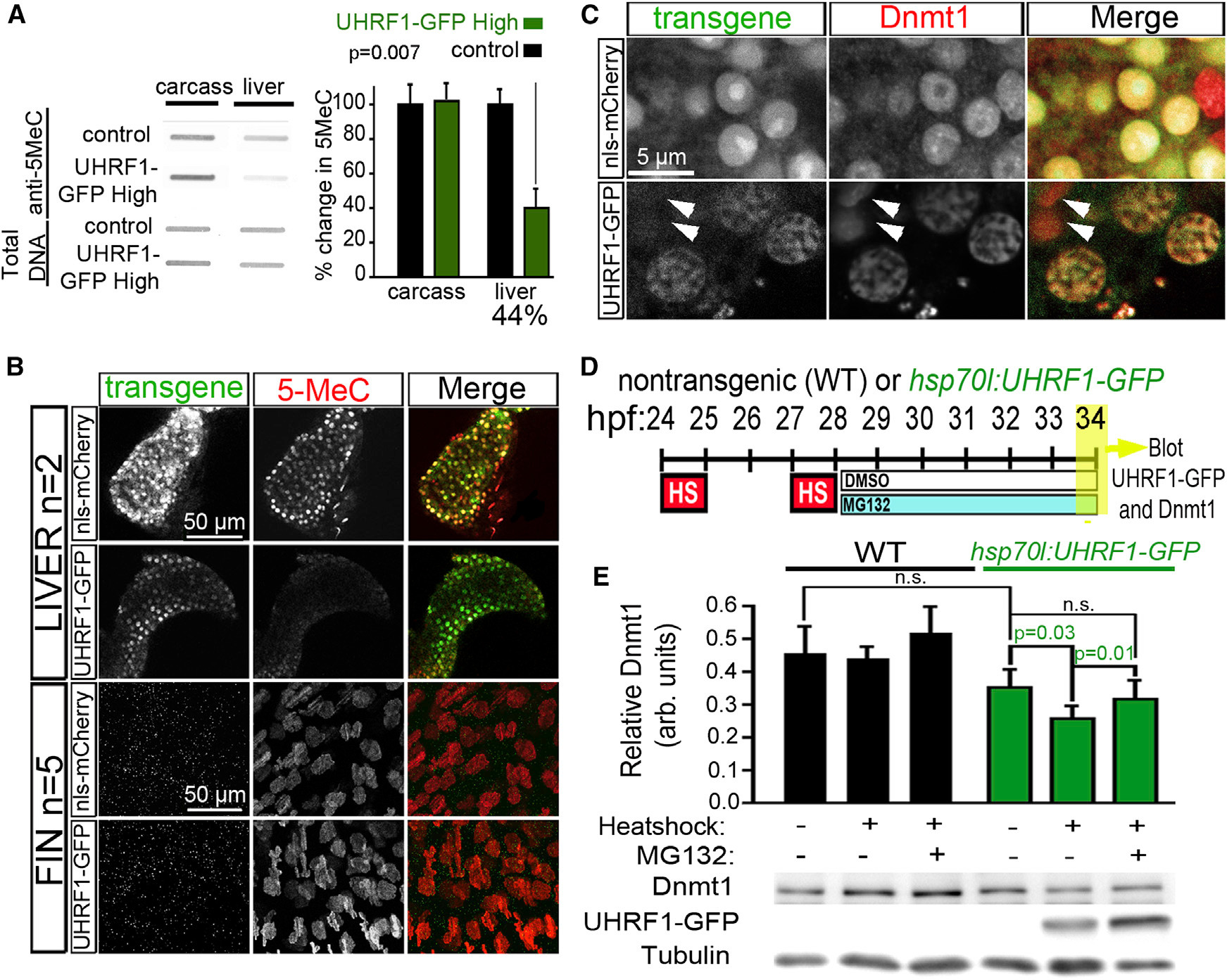
Fig. 1
High UHRF1 Expression Causes Global DNA Hypomethylation
(A) 5MeC levels and total DNA stained with methylene blue were measured in 5 dpf control and UHRF1-GFP High livers (n = 4) and liverless carcasses (n = 3). The ratio of 5MeC to total DNA was averaged and normalized to controls. Student’s t test was used to determine p values.
(B) Confocal stacks of livers (top) and fins (bottom) from 5 dpf nls-mCherry and UHRF1-GFP High larvae stained with anti-5MeC. Because a hepatocyte-specific promoter was used for transgenesis, there was no transgene expression in the fin. (C) Dnmt1 is uniform in the hepatocyte nucleus of four dpf nls-mCherry larvae but is found in GFP-containing punctae in UHRF1-GFP High hepatocytes. Arrows point to cells that do not express GFP and have Dnmt1 distribution pattern similar to controls.
(D) Tg(hsp70I:UHRF1-EGFP) and nontransgenic controls were heat shocked at 37C for 1 hr at 24 and 27 hpf, treated with 10 μM MG132 or DMSO at 28 hpf, and collected at 34 hpf for immunoblotting.
(E) Dnmt1 levels normalized to tubulin were averaged from six experiments. Student’s t test was used to determine p values; n.s., not significant; error bars represent SD.
Reprinted from Cancer Cell, 25(2), Mudbhary, R., Hoshida, Y., Chernyavskaya, Y., Jacob, V., Villanueva, A., Fiel, M.I., Chen, X., Kojima, K., Thung, S., Bronson, R.T., Lachenmayer, A., Revill, K., Alsinet, C., Sachidanandam, R., Desai, A., SenBanerjee, S., Ukomadu, C., Llovet, J.M., and Sadler, K.C., UHRF1 overexpression drives DNA hypomethylation and hepatocellular carcinoma, 196-209, Copyright (2014) with permission from Elsevier. Full text @ Cancer Cell