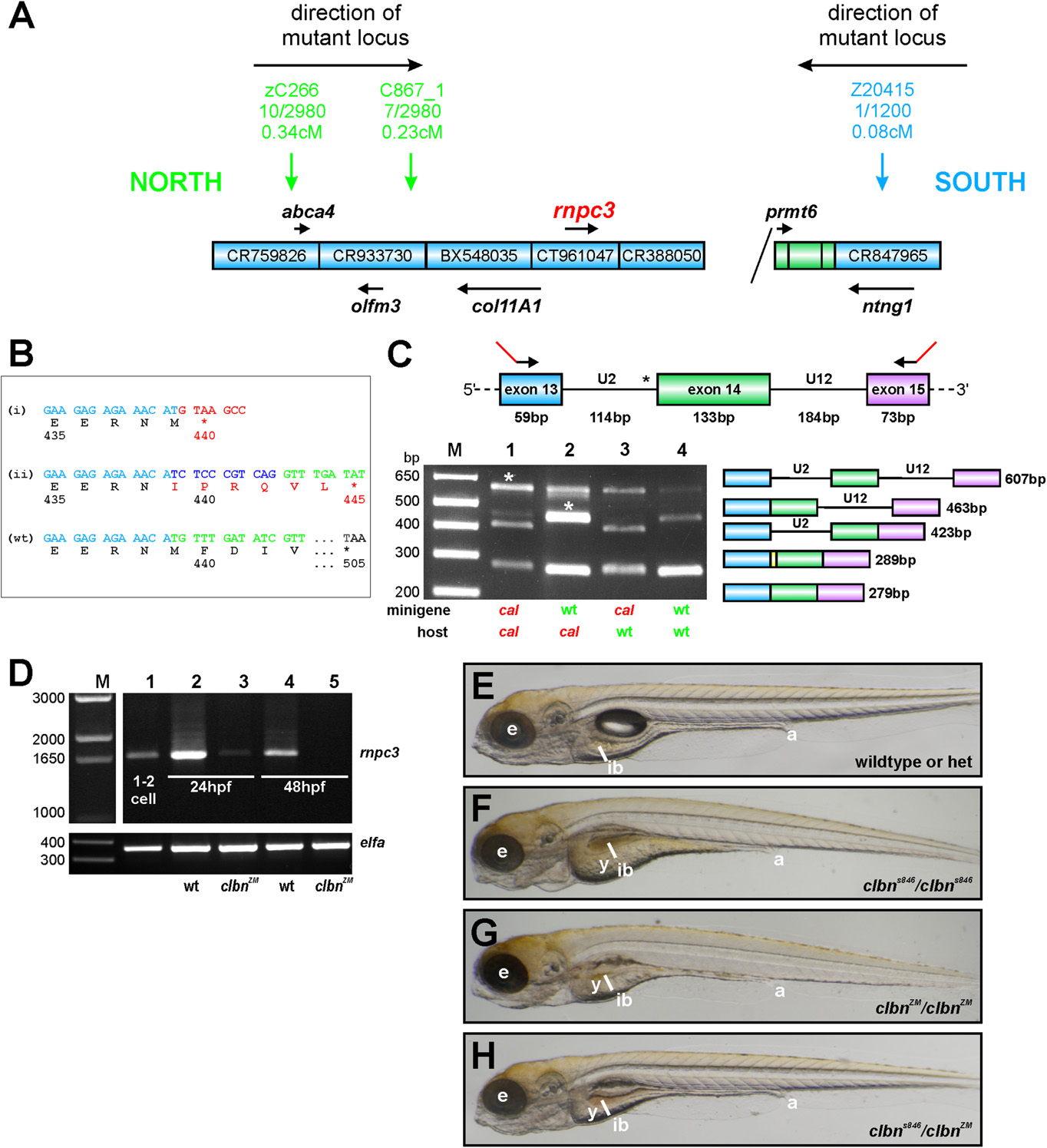
Fig. S1 rnpc3 is the mutated locus in clbn. (A) Simple schematic representation of the genetic interval containing the mutated locus in clbns486 based on the zebrafish genome assembly (Zv9). The genetic region of interest was defined by determining linkage of several simple sequence length polymorphism (SSLP) markers to the mutation (1). A total of up to 2,980 meioses from a defined series of crosses between the original ethylnitrosourea-mutagenized strain and a polymorphic mapping strain were analyzed, narrowing down the genetic interval containing the mutation to a distance of 0.31 cM between the closest linked markers. Blue boxes represent bacterial artificial chromosome (BAC) clones encompassing the region. Diagonal line indicates the presence of additional intervening sequence. Black arrows and gene names shown in italics represent annotated genes. (B) Aberrant transcripts extracted from clbns846 larvae encode truncated translation products. RT-PCR of rnpc3 RNA extracted from clbns846 larvae produces four faint bands (see Fig. 2A), corresponding to the aberrant transcripts (a–d) shown schematically in Fig. 2B. Product (i) is encoded by transcripts retaining intron 13 (a and c in Fig. 2B). A frame-shift in the coding sequence results in a premature stop codon instead of residue 440. Product (ii) is encoded by transcripts (b and d in Fig. 2B) using a de novo 32 splice site (ss) in intron 13 that results in the addition of 10 intronic nucleotides to the coding sequence. This also introduces a frame-shift and a premature stop codon is encountered instead of residue 445. Full-length WT product is shown below. (C) In vivo minigene assay to characterize the splicing defect in clbns846. A minigene spanning the U2-type intron 13, the U12-type intron 14, and flanking exons, was amplified from WT and clbns846 genomic DNA using primers carrying 52 tails (23 bp; shown in red) to allow specific amplification of the exogenous transgene after injection into zebrafish embryos. No correctly spliced RT-PCR product is detected upon injection of mutant minigene plasmid DNA into clbns846 and WT embryos (lanes 1 and 3, respectively). Sequencing of the lower bands confirms the use of the same de novo 32 ss detected endogenously in clbns846. The retention of 10 intronic nucleotides is indicated by the yellow region in the schematic diagram. Mutant larvae display significant retention of the U12-type intron 14 upon injection of both the WT and mutant minigene (lanes 1 and 2, asterisks). (D) Full-length rnpc3 cDNA is RT-PCR amplified from RNA extracted from one to two cell stage embryos (lane 1) indicating maternal deposits of WT rnpc3. The level of full-length rnpc3 is greatly reduced in clbnZM mutants by 24 hpf and undetectable by 48 hpf. Lanes contain RT-PCR products of RNA extracted from genotyped homozygous WT (lanes 2 and 4) and clbnZM mutant embryos (lanes 3 and 5) at 24 hpf (lanes 2 and 3) and 48 hpf (lanes 4 and 5). (Lower) elfa control. (E–H) Brightfield microscopy images of left lateral views of (E) WT siblings from a clbnZM × clbns846 cross, (F) homozygous mutant clbns846 larvae, and (G) homozygous mutant clbnZM larvae. (H) Twenty-five percent of offspring from clbnZM × clbns846 crosses show a phenotype indistinguishable from that of either clbns846 or clbnZM homozygous mutants. The images in panels E–H were taken at the same magnification.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Proc. Natl. Acad. Sci. USA