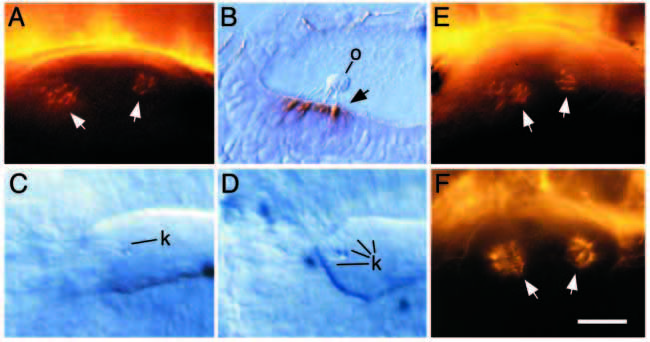
Fig. 4 Genetic interactions between noitb21, dlAdx2 and mibta52b. (A) Dorsolateral view of the otic vesicle of a 30 h noitb21/tb21 embryo stained with anti-Pax2 and anti-acetylated tubulin antibodies. Nuclear staining is not detectable, but the apical surfaces and kinocilia of hair cells are strongly labeled (arrows). Twice the normal number of hair cells are evident. (B) Parasagittal section of a noitb21/tb211 mutant stained in whole mount with anti-Pax2 and antiacetylated tubulin antibodies. Nuclear pax2 staining is not detectable, but hair cells are clearly marked by acetylated tubulin staining (arrow) and the close association with the otolith (o). (C) Otic vesicle of a live noitb21/tb21 mutant viewed under DIC optics at 19 h. The kinocilia (k) of two tether cells, and attached otolith precursors, are seen at the anterior end of the otic vesicle. (D) Otic vesicle of a live dlAdx2/dx2 mutant viewed under DIC optics at 19 h. The kinocilia (k) of supernumerary tether cells are evident. (E) Dorsolateral view of the otic vesicle of a 30 h dlAdx2/dx2; noitb21/tb21 double mutant stained with anti-Pax2 and anti-acetylated tubulin antibodies. Hair cells, detected by staining of their kinocilia and apical surfaces (arrows), are produced in numbers that are comparable to noitb21/tb21 single mutants. (F) Dorsolateral view of the otic vesicle of a 30 h mibta52b.ta52b; noitb21/tb21 double mutant stained with anti-Pax2 and anti-acetylated tubulin antibodies. Hair cells (arrows) are produced in much greater numbers than in noitb21/tb21 single mutants. In all panels, anterior is to the right and dorsal is up. Scale bar, 10 μm (C,D), 15 μm (B), or 25 μm (A,E,F).
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development