Fig. S1
G4C2 repeats expression vector cloning, related to Figure 1
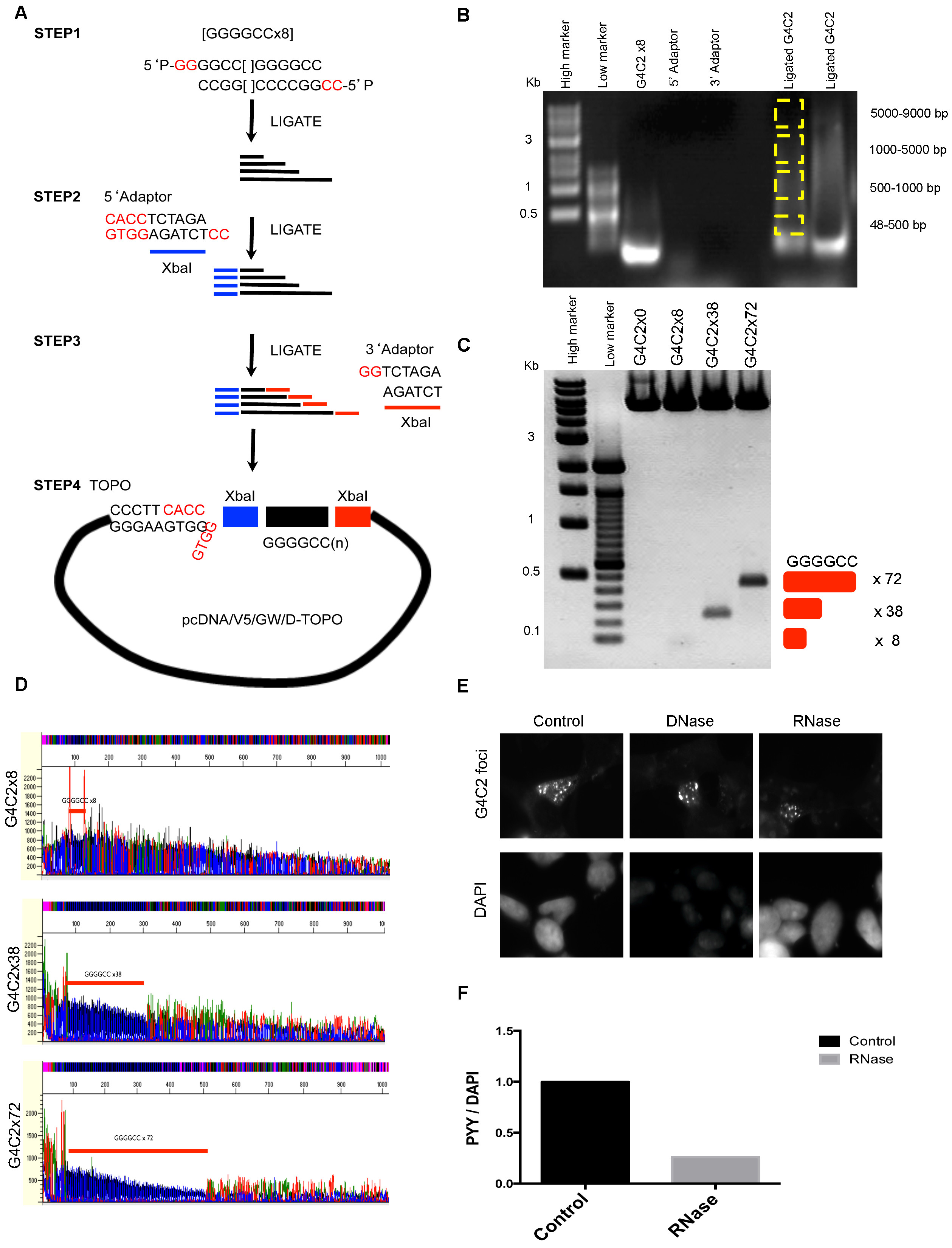
(A) Schematic diagram of G4C2 extension methods. 5′ phosphorylated oligo 5′P-GG- (G4C2)x8 and 5′P-CC-(C4G2)x8 form a sticky end with overhang of GG and CC after overnight ligation. (B) Ligation products were separated by gel electrophoresis and dissected according to the size of the G4C2 expansion (yellow box). Gel extracted DNAs were used for Topo cloning and positive colonies were selected by XbaI digestion. (C) Stable E.coli lines containing 8x, 38x, 72x were selected and the presence of the insert confirmed by XbaI digestion as indicated. (D) G4C2 DNA sequence were verified by dGTP secondary structure sequencing. G4C2 repeats were indicated with red bar. (E) SH-SY5Y cells were transfected with the G4C2x72 plasmid and G4C2 foci were detected after treatment with DNase (10 U/ml) or RNase (400 ng/ml) for 10 min at 37 °C. DAPI staining shows decreased intensity in DNase-treated cells. (F) To identify the level of degraded RNA, RNase-treated cells were stained with the RNA-specific dye Pyronin Y (PYY, 30 μg/ml) for 5 min at room temperature and staining intensity was measured using a FLUOstar plate reader (DAPI, 358 nm; PYY, 567 nm) and PYY values normalized to DAPI values.