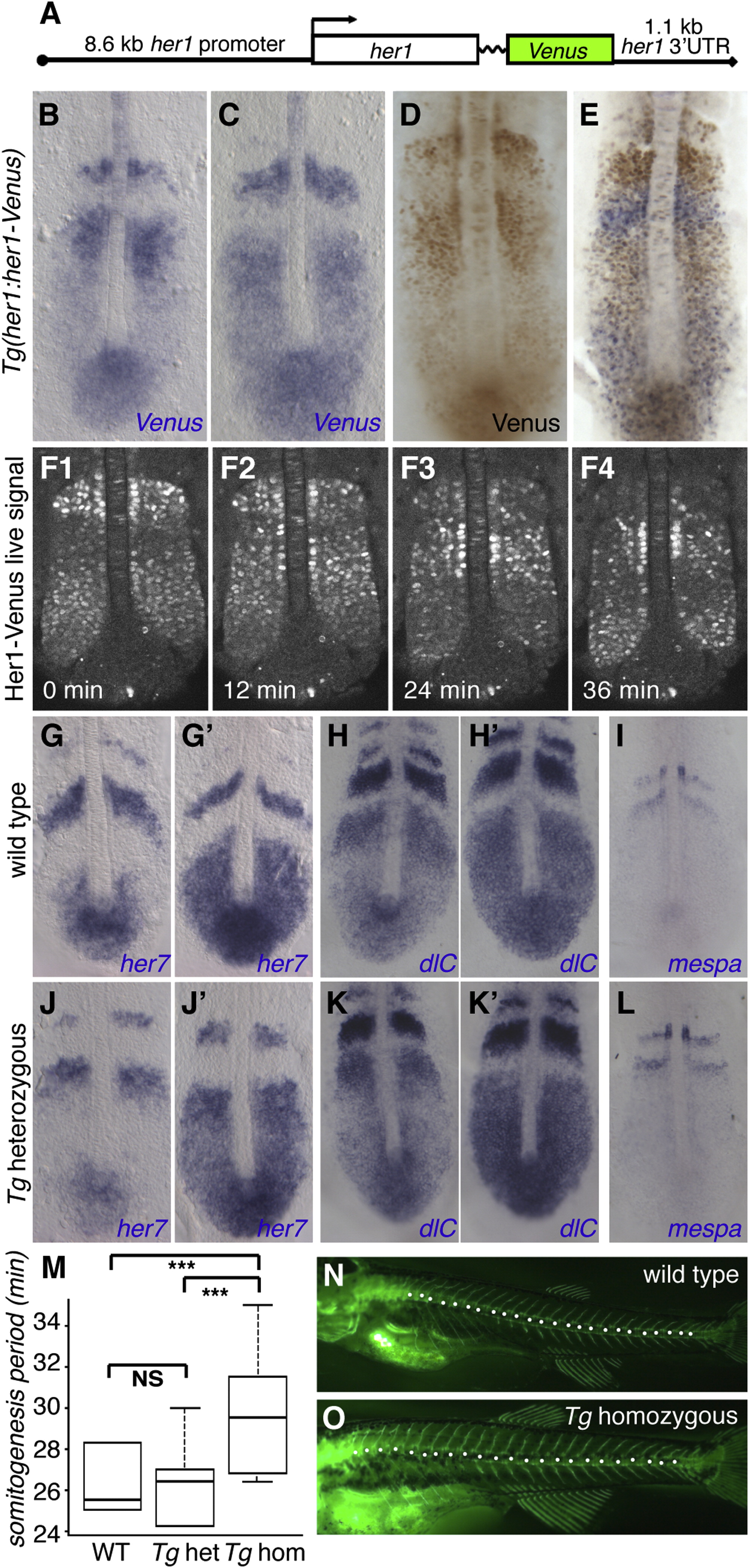
Fig. 1 The Zebrafish Transgenic her1:her1-Venus Line Recapitulates Dynamic her1 Expression(A) Diagram of the her1:her1-Venus construct.(B=E) Cyclic reporter transcript and protein expression in heterozygous Tg(her1:her1-Venus) embryos. Transcript (blue) and protein (brown) are largely expressed out of phase.(F) Still images from Movie S1 (in dorsal view) reveal waves of dynamic reporter expression in the PSM.(G-L) her7 (G, G′, J, and J′), dlc (H, H′, K, and K′), and mespa (I and L) expression in wild-type (G-I) and heterozygous Tg(her1:her1-Venus) (J-L) embryos.(M) Segmentation speed in wild-type (n = 20), her1:her1-Venus heterozygous (n = 26) and homozygous (n = 29) embryos. Somites were first counted at the two- to six-somite stage, and then again after 4-5.5 hr of development at 28°C. The box represents segmentation speed values between the first and third quartiles, the bold line represents the median value, and the whiskers represent maximal and minimal values. Homozygous transgenic embryos segmented significantly more slowly than heterozygote and wild-type embryos. A Wilcoxon rank sum test yielded significant differences for wild-type versus homozygote (p < 10-5), and heterozygote versus homozygote (p < 10-5), but not wild-type versus heterozygote (p = 0.87) embryos.(N and O) Segment number and size in wild-type (H) or homozygous her1:her1-Venus (I) larvae. Calcified skeletal structures were revealed at 21 dpf by calcein staining (Du et al., 2001). The total number of precaudal, transitional, and caudal segments (white dots; Bird and Mabee, 2003) was significantly lower in homozygous transgenic larvae (median = 21 segments, n = 64) than in wild-type larvae (median = 24, n = 33; Wilcoxon rank sum test, p < 10-5). Note that the segment size appears larger in homozygous her1:her1-Venus compared with wild-type larvae.See also Figures S1 and S2 and Movie S1.
Reprinted from Developmental Cell, 23(5), Delaune, E.A., François, P., Shih, N.P., and Amacher, S.L., Single-cell-resolution imaging of the impact of notch signaling and mitosis on segmentation clock dynamics, 995-1005, Copyright (2012) with permission from Elsevier. Full text @ Dev. Cell