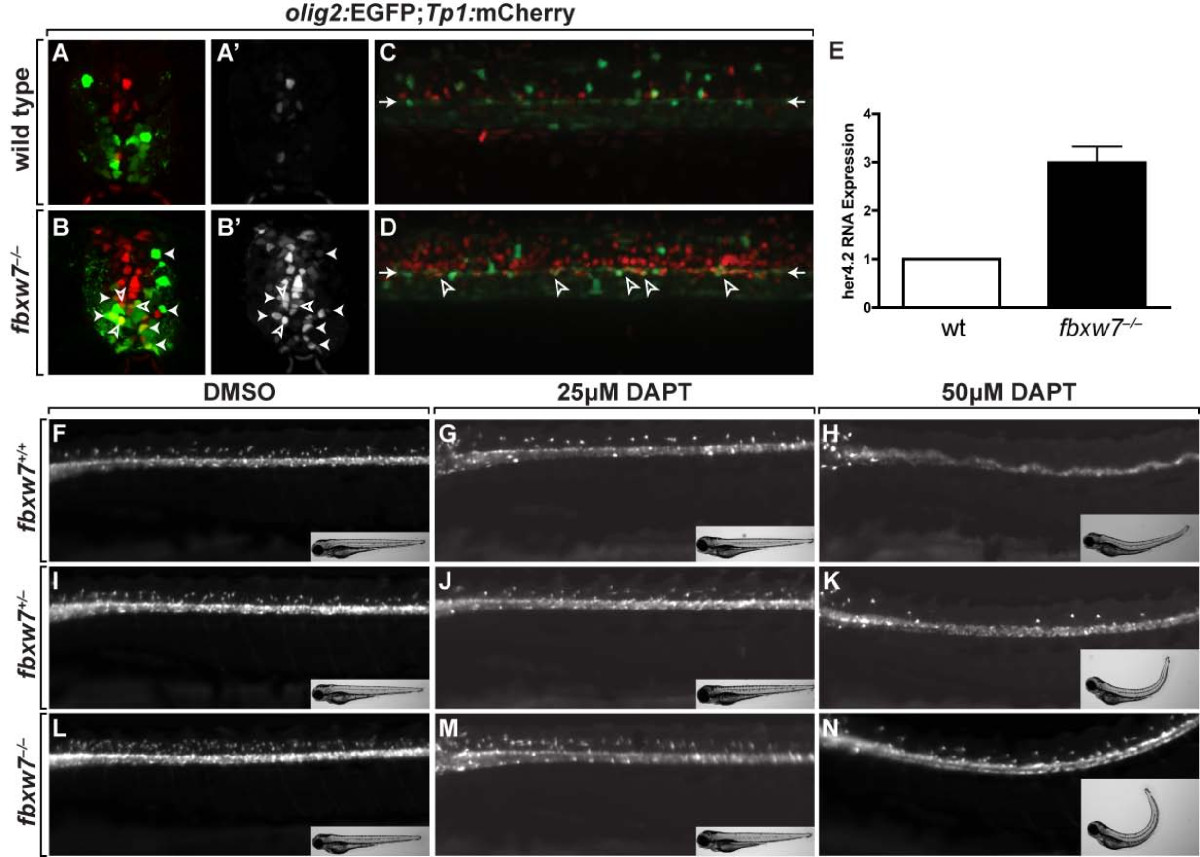
Fig. 5 Fbxw7 regulates oligodendrocyte number by regulating Notch signaling. (A,B) Transverse sections of 3 dpf wild-type (A) and fbxw7vu56 mutant (B) spinal cords showing Tp1:mCherry and olig2:EGFP expression. Outlined arrowheads mark Tp1:mCherry+, olig2:EGFP+ precursors. Solid arrowheads indicate Tp1:mCherry+, olig2:EGFP+ oligodendrocyte lineage cells. (A′,B′) Tp1:mCherry images alone. (C,D) Confocal z stack projections, from lateral view, of olig2:EGFP and Tp1:mCherry in 6 dpf wild-type sibling and fbxw7vu56 mutant larvae. Arrowheads point to row of olig2:EGFP+ precursors lining the central canal. Outlined arrowheads mark Tp1:mCherry+, olig2:EGFP+ precursors. (E) Relative levels of her4.2 mRNA in 4 dpf wild-type and fbxw7vu56 mutant larvae determined by quantitative PCR. Wild-type level was assigned an arbitrary level of 1.0 (p = 0.0272). (F-N) Lateral images of living 3 dpf larvae treated with DMSO or DAPT plus DMSO from 36–48 hpf. All larvae carried the Tg(olig2:EGFP) reporter to mark OPCs and oligodendrocytes in dorsal spinal cord. DAPT reduced the number of oligodendrocyte lineage cells in larvae of each genotype. Insets show bright field images of larvae. Each larva was genotyped following the experiment.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Neural Dev.