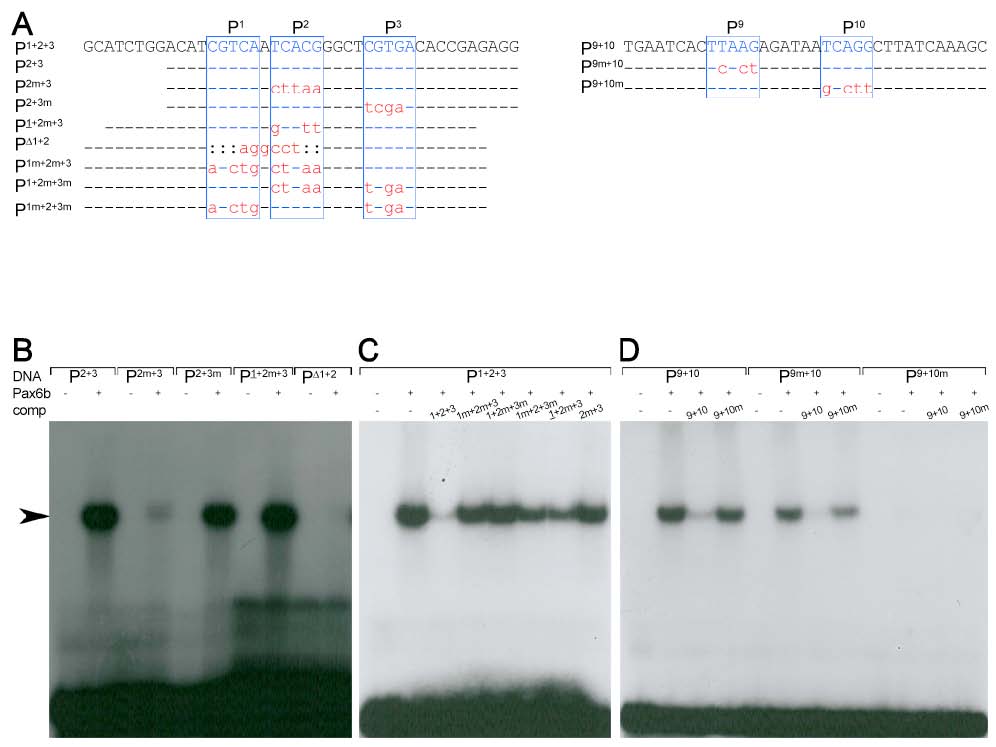
Fig. S3
Pax6b binding properties of the sequence motives P1-3 and P9-10
(A)Sequence of oligonucleotides used in the EMSA studies (top row). Indicated are the Pax6 core consensus sequences P1, P2, P3, P9 and P10 (blue box, letters in upper case) and nucleotides that were unchanged (line), mutated (red letters in lower case) or deleted (colon) in comparison to the genomic sequence. (B-D) Radiograms of EMSA studies with corresponding 32P-labeled oligonucleotides (DNA, as indicated), Pax6b protein and nonlabelled competitor DNA (comp, as indicated). (B) Weak or missing signals (arrow) for P2m+3, PΔ1+2 and strong signals for P2+3, P2+3m and P1+2m+3 suggest that Pax6b binds with high affinity to P1 and P2 but not to P3. (C) The inefficient competition of all mutated oligonucleotides in comparison to P1+2+3 shows that highest affinity binding requires the presence of both sites, P1 and P2. (D) Missing signals for and inefficient competition by P9+10m show that Pax6b binds with high affinity to P10 but not to P9.
Reprinted from Developmental Biology, 365(1), Arkhipova, V., Wendik, B., Devos, N., Ek, O., Peers, B., and Meyer, D., Characterization and regulation of the hb9/mnx1 beta-cell progenitor specific enhancer in zebrafish, 290-302, Copyright (2012) with permission from Elsevier. Full text @ Dev. Biol.