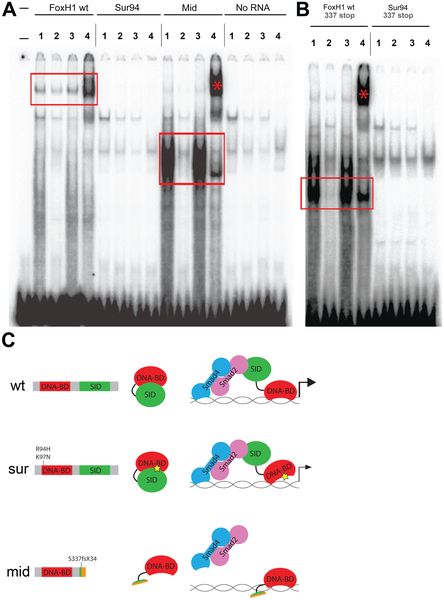
Fig. 4
DNA–binding activities of FoxH1 mutants.
(A) Electrophoretic mobility shift assays using a FoxH1 binding site probe derived from the zebrafish gsc promoter and in vitro-translated epitope-tagged full-length proteins. FoxH1 and Mid protein bind the probe in a protein- and sequence-specific manner (red boxes and asterisk), while Sur protein shows no binding activity. Upper lane labels indicate the RNA translated for use in each binding reaction; individual lane numbers denote additions to the basic binding reactions (1: no additions; 2: 100-fold excess unlabeled competitor probe; 3: 100-fold excess mutated unlabeled competitor probe; 4: anti-HA antibody). (B) EMSAs using truncated FoxH1 and Sur proteins lacking the C-terminal SID. Truncated wild-type protein specifically binds the probe (red box), while truncated Sur protein does not. Lane markings are as described in (A) above. (C) Model for DNA-binding activities of wild-type and mutant FoxH1 proteins. Wild-type protein binds weakly to its recognition sites alone, but can bind strongly upon loss of its C-terminus, suggesting that Smad interaction may “open up” the conformation of the wild-type protein and allow for strong binding upon pathway activation. Sur protein is impaired in its DNA-binding ability, but may be weakly/transiently tethered to its recognition sequences by activated Smads or other unknown factors. Mid protein cannot interact with Smads and so cannot transduce Nodal signals, but can bind strongly to FoxH1 recognition sites.