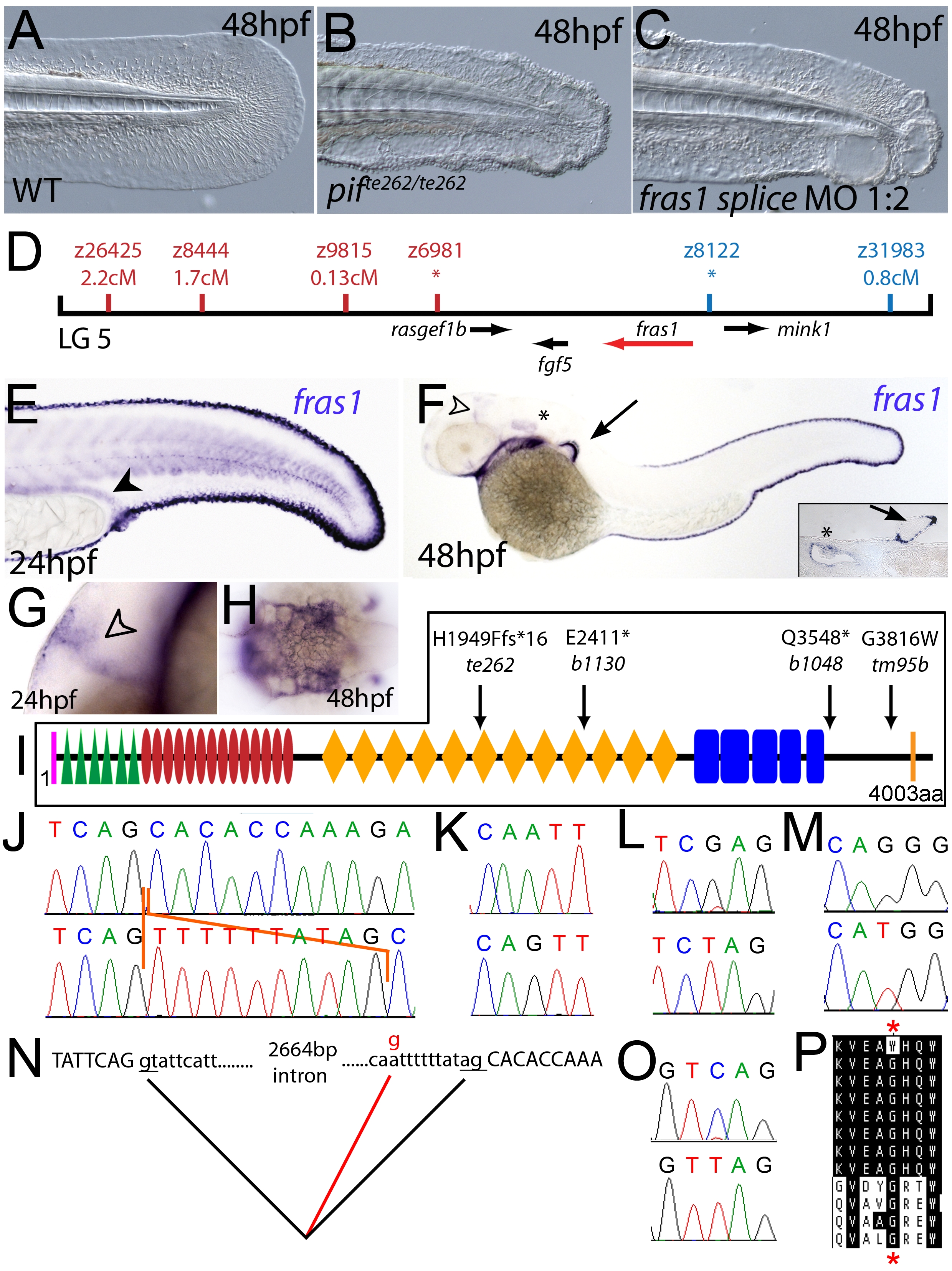
Fig. 2 The fin blistering of pinfin mutants is caused by mutations in Fras1.
(A–C) Injection of a morpholino targeting the translation site of fras1 mRNA into wild-type embryos phenocopies the pif fin blisters; lateral views of tip of tail at 48 hpf; (A) uninjected control; (B) pifte262/te262 mutant; (C) fras1 morphant. (D) Linkage analysis localised the pinfin locus to linkage group 5, close to the SSLP markers z6981 and z8122, neither of which recombined with the locus, and between which the fras1 gene (depicted in red) was located. (E–H) Images of embryos stained by in situ hybridisation using a probe against fras1. Close up view of the tail region of a 24 hpf embryo (E) and a 48 hpf embryo (F) are shown. The fras1 gene is expressed in the medial fin fold, the pectoral fin fold (arrow in F; also see inset), the pronephric ducts (filled arrowhead in E) and the otic vesicle (asterisks in F and insert). fras1 expression is also noted in the midbrain-hindbrain region at 24 hpf and 48 hpf (open arrowhead in G and F) and in the pharyngeal pouches of the arches (H). (E–G) show lateral views, (H) a dorsal view. (I) Schematic of the predicted domain structure of zebrafish Fras1 protein, with a signal peptide (pink bar), von Willebrand C domains (green triangle), furin-like domains (red ovals), CSPG domains (orange diamonds), Calx-bdomains (blue boxes) and a transmembrane domain (orange bar). Lesions found in four pif alleles are indicated above the protein schematic. (J,L,M,O) Sequence chromatograms of fras1 cDNA from pifte262/te262 (J), pifb1130/b1130 (L), pifb1048/b1048 (O), and piftm95/tm95 (M) mutants, displaying the lesions depicted in (I). The frame shift (fs) mutation in pifte262 is due to the insertion of the last 10 nucleotides of intron 42 (delineated with orange lines). In all panels, the WT sibling chromatogram is given above the mutant chromatogram. (K) Chromatograms showing the genomic sequence at the end of intron 42 of the fras1 gene in pifte262/te262 (lower panel) and WT sibling (upper panel) embryos. The A>G substitution generates a novel splice acceptor. (N) Representation of the exon 42-exon 43 junction with the mutation generating aberrant splicing (red line). (P) The glycine residue (red asterisks) substituted in piftm95/tm95 is strictly conserved in both Fras1 and Frem2 proteins across diverse phyla, as seen in an alignment of the region. Protein sequences are from top: zebrafish piftm95/tm95 mutant Fras1; zebrafish wild-type Fras1; fugu Fras1; human FRAS1; mouse Fras1; dog Fras1; cow Fras1; chicken Fras1; sea urchin ECM3; zebrafish Frem2a; fugu Frem2a; human FREM2.