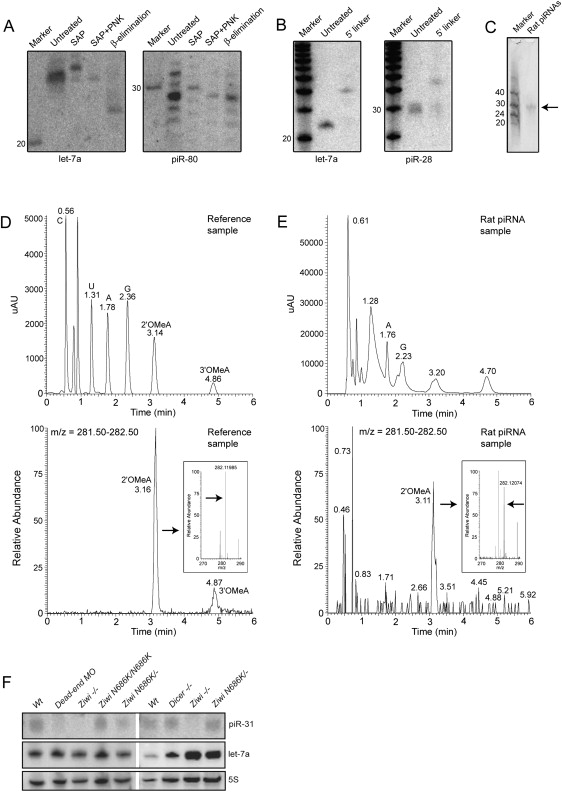
Fig. 6 Biochemical and Genetic Analysis of piRNAs
(A) Left panel shows a northern blot for let-7a. Samples are treated with Shrimp Alkaline Phosphatase (SAP), SAP+ polynucleotide kinase (PNK), and NaIO4 oxidation followed by β-elimination. The right panel shows the same blot probed for piR-80.
(B) Ligation of a 17 nt linker with both 5′ and 3′ hydroxyl groups to both let-7a and piR-28 is shown.
(C) Purified rat piRNA is shown. Approximately 0.3% of the purified product was run on a gel and stained with SYBRGold.
(D) LC/MS characterization of a mix of synthetic C, A, U, G, 2′-OMeC, 3′-OMeC, 2′-OMeA, and 3′-OMeA is shown. In the top graph UV intensity is plotted versus time. In the lower graph the presence of molecules with an m/z in the range of OMeA (m/z = 282.12) is plotted versus time, revealing peaks at the retention times (RT) of 2′ and 3′-OMeA. The inset shows high resolution m/z determination (RT = 3.06–3.25), with an arrow pointed at the exact mass of 2′-OMeA. Numbers in the graph indicate RT of indicated peak.
(E) Similar experiment as in (D) but now with nucleosides derived from rat piRNAs. Two UV peaks are observed around the RT of 2′ and 3′-OMeA. The peak overlapping with 2′-OMeA contains molecules with the mass of OMeA (m/z = 282.12 at RT = 3.05-3.22; lower graph and inset). This species is present in the left shoulder of the UV peak at RT = 3.20. Note that most of the UV signal of the peak at RT = 3.20 does not come from 2′OMeA. The UV peak around the RT of 3′-OMeA contains no species with a molecular weight of 3′-OMeA (not shown). Coinjection with synthetic 2′ and 3′-OMeA confirms these results (Figure S7).
(F) Northern blot analysis of piRNA expression is shown in various mutant backgrounds. Upper panel shows a northern blot for piRNA piR-31. The same blot was probed for miRNA let-7a (middle panel) and 5S RNA (lower panel). Note that let-7a is expressed in the somatic germline (not shown), so disruption of dicer in the germ cells has no effect on let-7a processing.
Reprinted from Cell, 129(1), Houwing, S., Kamminga, L.M., Berezikov, E., Cronembold, D., Girard, A., van den Elst, H., Filippov, D.V., Blaser, H., Raz, E., Moens, C.B., Plasterk, R.H., Hannon, G.J., Draper, B.W., and Ketting, R.F., A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish, 69-82, Copyright (2007) with permission from Elsevier. Full text @ Cell