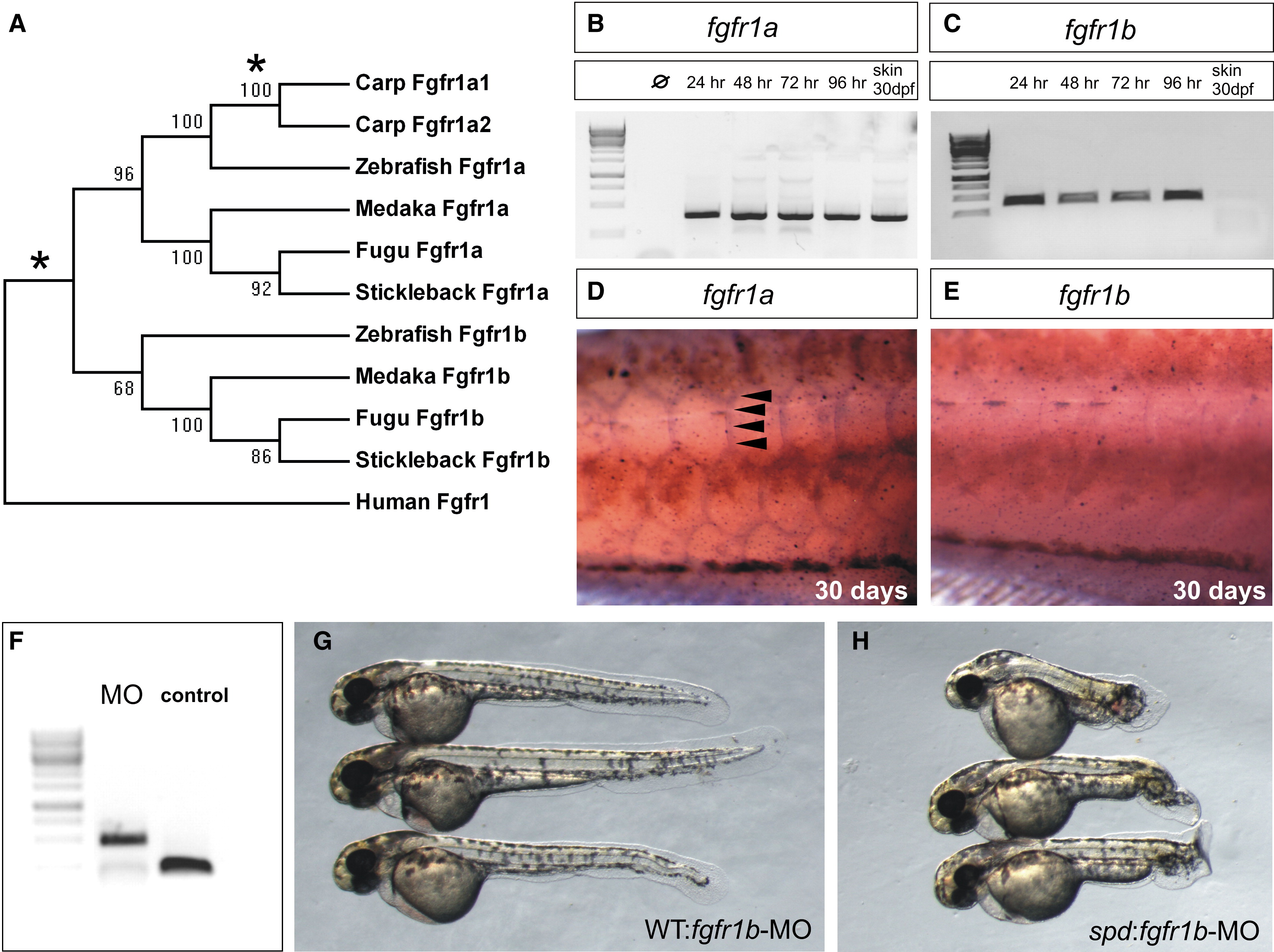
Fig. 3 Specific Postembryonic Function of the Zebrafish fgfr1a Gene and Functional Redundancy of fgfr1 Paralogs during Early Embryogenesis
(A) Neighbor-joining consensus tree of fgfr1 paralogs in teleosts. Bootstrap values are listed at nodes. Human Fgfr1 is shown as a representative of the sarcopterygian clade. Asterisks denote positions of whole-genome duplication events.
(B and C) Analysis of the expression of zebrafish fgfr1a (B) and fgfr1b (C) paralogs during development shows expression through early development of both genes by RT-PCR (35 cycles). Only fgfr1a expression was detected in RNA of skin from fish at 30 days postfertilization (dpf).
(D and E) In situ hybridization analysis shows the differential expression of the two paralogs during scale formation. Arrowheads point to regions of expression.
(F) Morpholinos (MO) targeting splice site junctions of fgfr1b effectively blocked splicing, leading to inclusion of an intronic sequence assayed by PCR amplification of cDNA.
(G and H) Injection of fgfr1b morpholino into wild-type embryos (G) shows little effect, whereas injection into spd embryos (H) results in specific posterior mesenchyme deficiencies of the tail.