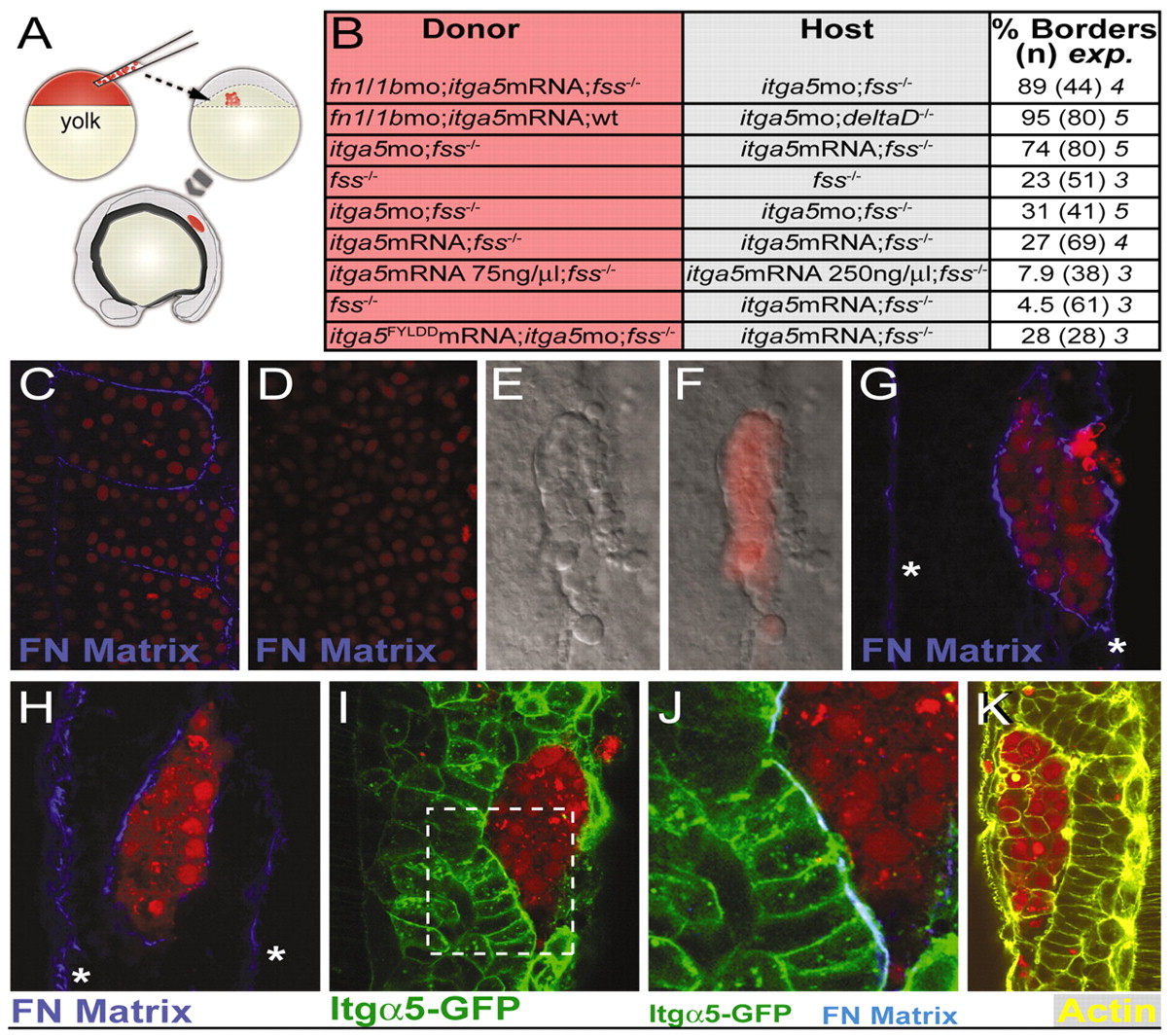
Fig. 4 Itgα5 non-cell-autonomously inhibits clustering and FN matrix assembly. (A) Genetic mosaics were made by transplanting labeled cells (red) from the blastula of a donor zebrafish embryo into the late blastula of an unlabeled host embryo. The chimeras were later analyzed for morphological border formation, greater than three cell diameters in length, around clones within the paraxial mesoderm. (B) Summary of the genetic mosaic experiments. Donor and host genotypes and treatments are indicated. The percentage of clones with borders, the total number of embryos examined and the number of experiments are displayed. (C,D) FN matrix (C) is eliminated by injection of fn1/fn1b morpholinos (D). FN matrix is blue and nuclei are red. (E,F) DIC image (E) and composite of a clone (F, red) with a border separating it from the host cells. (G) FN matrix forms along a clone lacking fn but expressing itga5 within a host lacking itga5 but expressing fn. Asterisks indicate FN matrix along the medial and lateral surfaces of the paraxial mesoderm. (H) FN matrix forms along a clone lacking itga5 within a host expressing itga5. (I) In mosaics of the same genotype as in H, Itgα5-GFP clusters along the clone. (J) The boxed region in I shown with FN matrix (blue) colocalizing with the clustered Itgα5-GFP. Note the columnar morphology of the host border cells. (K) Actin belts are seen along the clone borders. (C-K) Anterior is up.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development