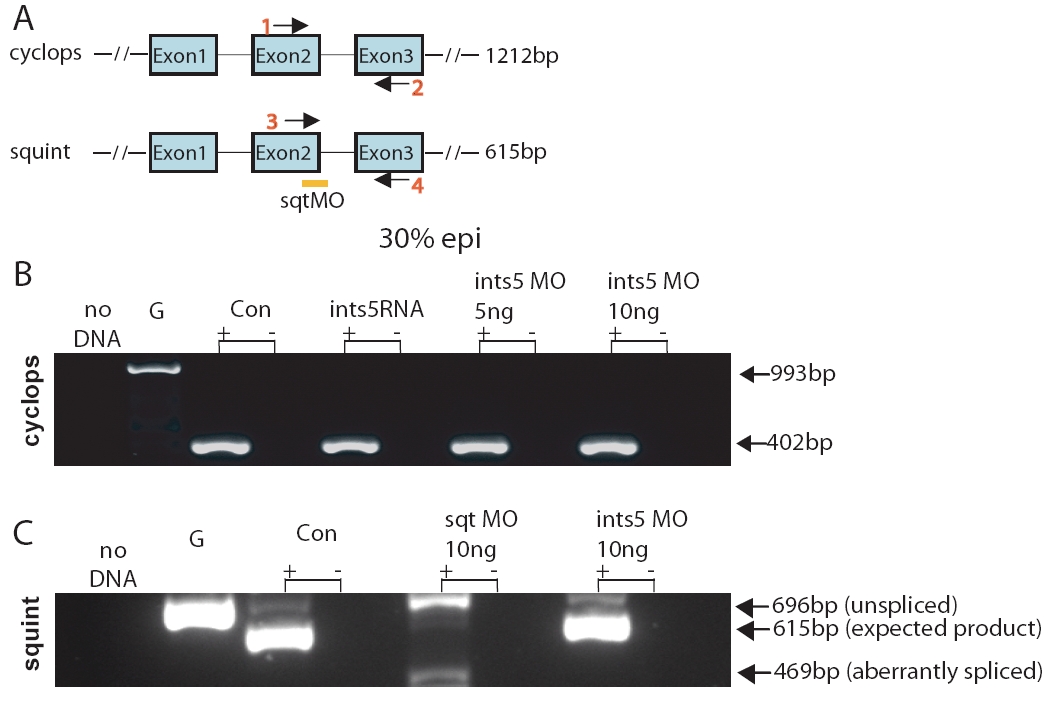
Fig. S5 Knockdown of Ints5 does not affect splicing of cyclops and squint RNA. (A) Schematic representation of cyclops and squint genomic loci. Arrows indicate the position of primer pairs used in RT-PCRs to detect splicing. Orange bar indicates the position of the squint splice junction morpholino at the exon2-intron2 boundary (Gore et al., 2005). Expected sizes of the amplified products are shown on the right. At 30% epiboly, a 402 bp cyc product is detected from all cDNA templates (B), whereas a 993 bp band is amplified from genomic DNA. (C) Embryos injected with ints5 morpholinos or control morpholinos show the expected 615 bp squint product. By contrast, embryos injected with squint splice-junction morpholinos show either unspliced (696 bp) or aberrantly spliced (469 bp) squint transcripts.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development