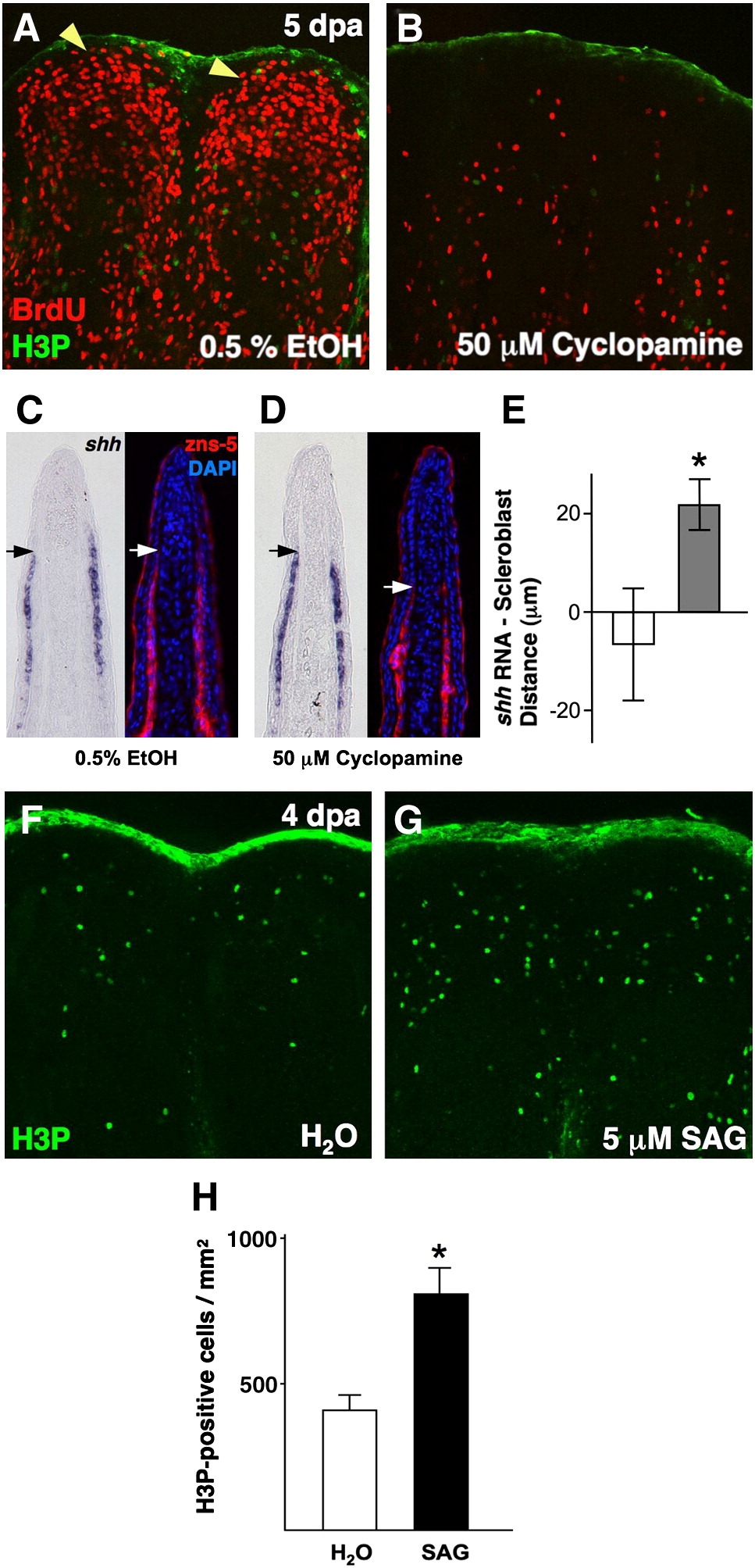
Fig. 3 Pharmacologic manipulation of Hh signaling in ongoing regenerates affects blastemal proliferation and scleroblast patterning. (A, B) Analysis of BrdU incorporation in 5 dpa fin regenerates (26 °C) of animals treated with 50 μM cyclopamine for 28 h. Cyclopamine treatment dramatically reduced blastemal BrdU incorporation (B), as compared to vehicle-treated control animals (A; yellow arrowheads indicate blastema). (C, D) Serial sections of vehicle- (C) and cyclopamine-treated fin regenerates (D) stained for shh mRNA localization or scleroblasts (zns-5). Mesenchymal scleroblasts align with epidermal shh signals in vehicle-treated regenerates, while they significantly trail these signals after 28 h cyclopamine treatment. Arrows indicate the distal limit of detectable shh or scleroblasts. (E) Quantification of distance between distal limits of shh expression and patterned scleroblasts. White bar = vehicle; gray bar = cyclopamine (n = 11; mean ± SEM; Student′s t-test, *P < 0.05). (F, G) Images of H3P staining of 4 dpa fin regenerates (26 °C) of animals treated with 5 μM SAG or vehicle for 2 h. Activation of Hh signaling by SAG treatment enhanced the number of mitotic blastemal cells (G). (H) Quantification of mitotic counts in SAG- and vehicle-treated animals. White bar = vehicle; black bar = SAG (n = 12; mean ± SEM; Student′s t-test, *P < 0.05).
Reprinted from Developmental Biology, 331(2), Lee, Y., Hami, D., De Val, S., Kagermeier-Schenk, B., Wills, A.A., Black, B.L., Weidinger, G., and Poss, K.D., Maintenance of blastemal proliferation by functionally diverse epidermis in regenerating zebrafish fins, 270-280, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.