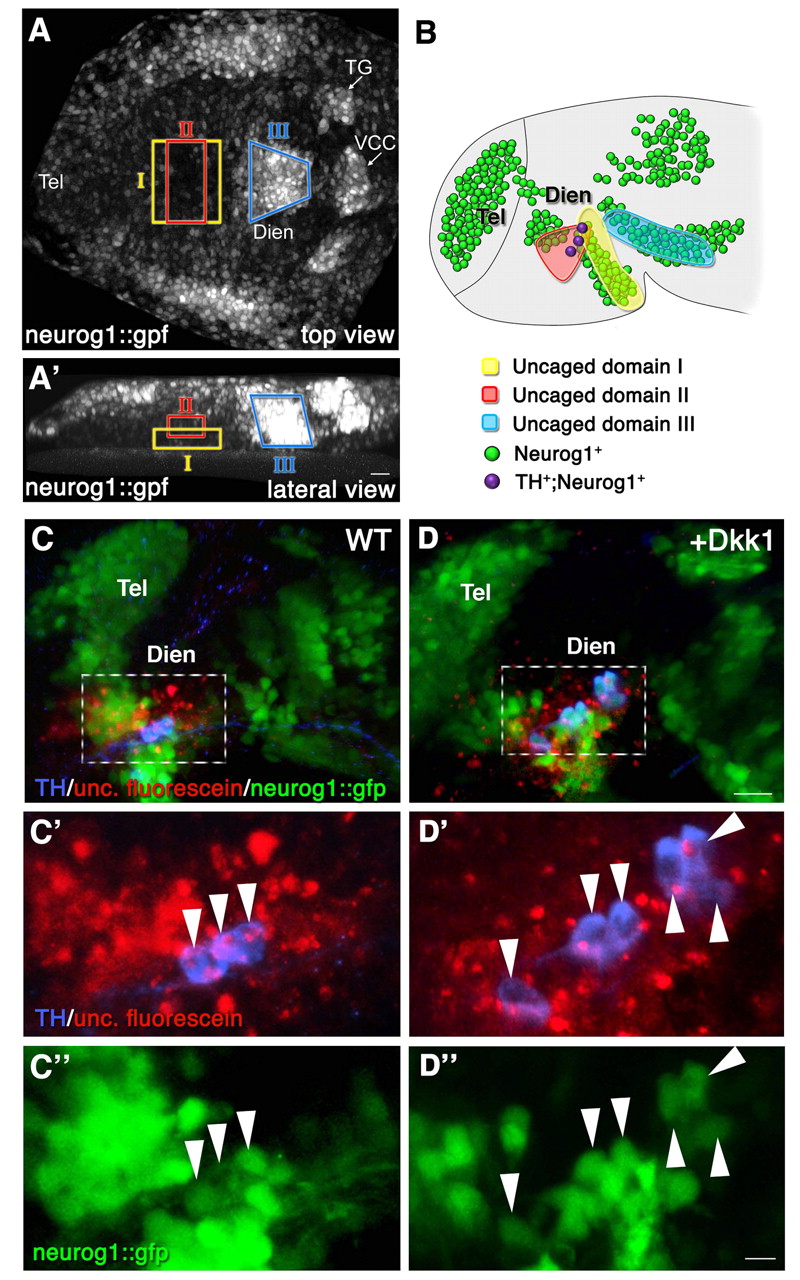
Fig. 3 Fate mapping of the diencephalic progenitor zones. High-resolution fate mapping by two-photon-based uncaging procedure. (A,A') Zebrafish embryos (anterior to the left) expressing GFP under the control of the neurog1 promoter (neurog1::gfp) were injected with caged dextran-fluorescein tracer dye at the 1-cell stage. At the 1- to 3-somite stage, the dye was uncaged at the indicated domains (denoted I, II and III) of the diencephalic anlage. (A) Dorsal view; (A′) lateral view. (B) Schematic summarizing the results of multiple uncaging experiments showing the final destination of the fluorescein-labeled cells in 24-hpf neurog1::gfp embryos. The clones corresponding to each of the uncaged domains are color coded (I, n=6; II, n=6; III, n=10). (C-D″) Control (WT; C-C″) and dkk1 mRNA-injected (D-D″) embryos that underwent uncaging were fixed at 24 hpf, followed by immunofluorescence staining of the uncaged fluorescein and of TH+ DA neurons. High-magnification images of a diencephalic area (dashed boxes in C,D) containing neurog1+ TH+ fluorescein+ triple-positive cells (arrowheads) are shown in C′,C″,D′,D″. Dien, diencephalon; Tel, telencephalon; TG, trigeminal ganglion; VCC, ventrocaudal cluster. Scale bars: 25 μm in A-D; 50 μm in C′,C″,D′,D″.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development