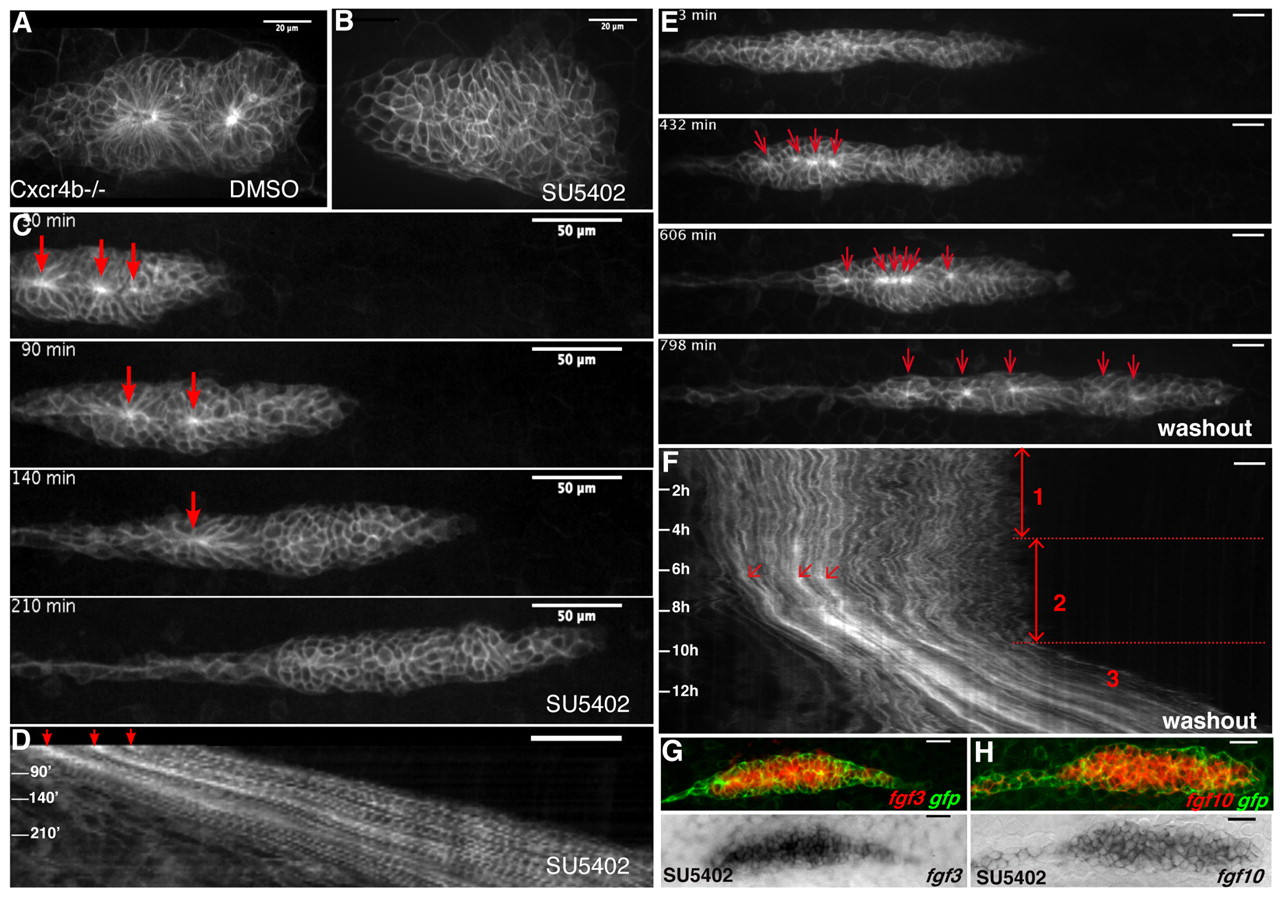
Fig. 5 FGF is required for rosette assembly, a prerequisite for primordium migration. Primordia of cxcr4b mutant embryos treated with DMSO (A) or SU5402 (B), showing the complete disappearance of rosettes after blocking the FGF pathway for 6 hours. (C,D) Time-lapse analysis and corresponding kymograph showing the progressive melting of rosettes after treatment with 80 μM SU5402. The migration speed is normal (70 μm/hour) and remains constant during this phase. Scale bar: 50 μm. (E,F) Time-lapse analysis and corresponding kymograph of a washout experiment showing that five rosettes (red arrows in E,F) simultaneously reassemble before migration resumes. Three phases can be distinguished on the kymograph: (1) uncoordinated migration, (2) rosette reassembly, and (3) migration recovery. (G,H) Expression of Fgf ligands in cldnb:gfp embryos treated for 6 hours with SU5402. Lower panels are transmission images of NBT/BCIP stainings. fgf3 (G) and fgf10 (H) are strongly expressed throughout the primordium when Fgfr is blocked. Scale bars: 20 μm (unless stated otherwise).
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development