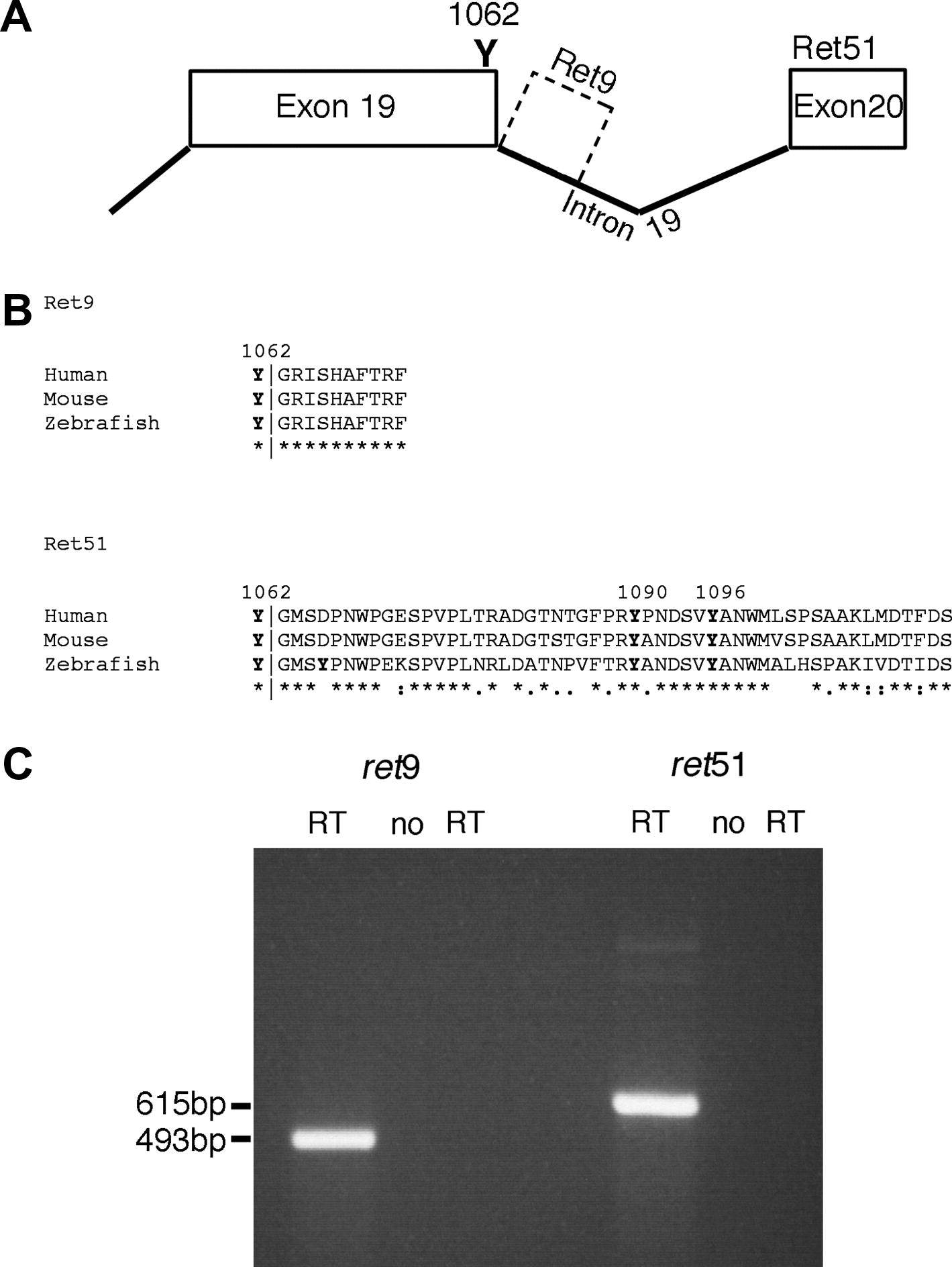
Fig. 1 Zebrafish ret exists as ret9 and ret51 isoforms. (A) Analysis of genomic sequences reveals that in zebrafish, alternative splicing at exon 19 can lead to generation of two isoforms of ret. Splicing to exon 20 generates ret51, whereas an absence of splicing generates ret9 from intronic sequences. (B) Sequence alignment using ClustalW 1.83 shows that zebrafish Ret9 amino acid sequences are identical to mouse and human RET9, and that zebrafish Ret51 sequences show significant sequence homology to mouse and human RET51, with 67% amino acid identity to each, including tyrosines (Y) at positions 1090, and 1096. Y1062, which is present in both Ret9 and Ret51, is also conserved. In alignment, “*” indicates identical amino acids, “:” indicates conserved substitutions, and “.” indicates semi-conserved substitutions (C) RT-PCR analysis of 24hpf zebrafish embryos using primers specific to ret9 and ret51 reveals that both isoforms are produced. These primers show no contaminating genomic DNA amplification in the “no RT” controls.
Reprinted from Mechanisms of Development, 125(8), Heanue, T.A., and Pachnis, V., Ret isoform function and marker gene expression in the enteric nervous system is conserved across diverse vertebrate species, 687-699, Copyright (2008) with permission from Elsevier. Full text @ Mech. Dev.