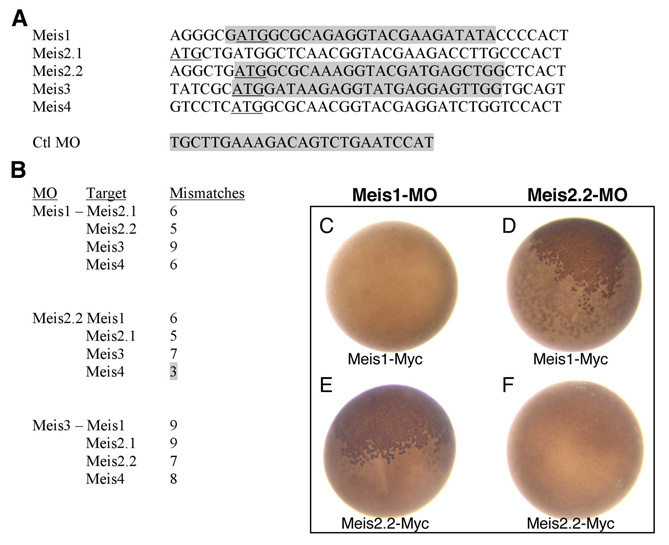
Fig. S1 Morpholino (MO) sequences, targets and specificity assay in Xenopus. (A) Alignment of Meis sequences around the ATG (underlined). Morpholino (targeted) sequences are shadowed in gray. The control mopholino used in this paper (Ctrl-MO) was originally designed to target the ATG region of Xenopus tropicalis Olig2. It does not target any ATG in the zebrafish genome. (B) Mismatches between meis1, meis2.2 and meis3 morpholinos and the ATG regions of non-target Meis genes. All but one have five or more mismatches, which should prevent cross-targeting. The exception is meis2.2-MO, which might cross-target meis4. This fact did not interfere in our analysis since meis4 is not expressed in the developing eye. meis3-MO, which has nine and seven mismatches with meis1 and meis2.2, respectively, is included here as it also served as control MO in some experiments. (C-F) Specificity assay in Xenopus laevis embryos. meis1 and meis2.2 mRNAs, carrying a C-terminal Myc tag (meis1-Myc and meis2.2-Myc) (10 ng), were co-injected with either meis1 or meis2.2 MOs (1 ng) in two-celled Xenopus embryos. After injection, embryos (n>50 per experiment) were allowed to develop for 24 hours at 18°C, fixed and immunostained to detect the Myc tag (e.g. Tena et al., 2007). Effective targeting results in the block of mRNA translation and no production of the Myc-tagged protein. Each MO blocks the translation of only its specific mRNA (C,F; absence of nuclear Myc staining). Accordingly, meis1-MO is unable to block meis2.2-Myc translation, as meis2.2-MO is incapable of blocking that of meis1-Myc (B,E; presence of nuclear Myc staining, brown).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development