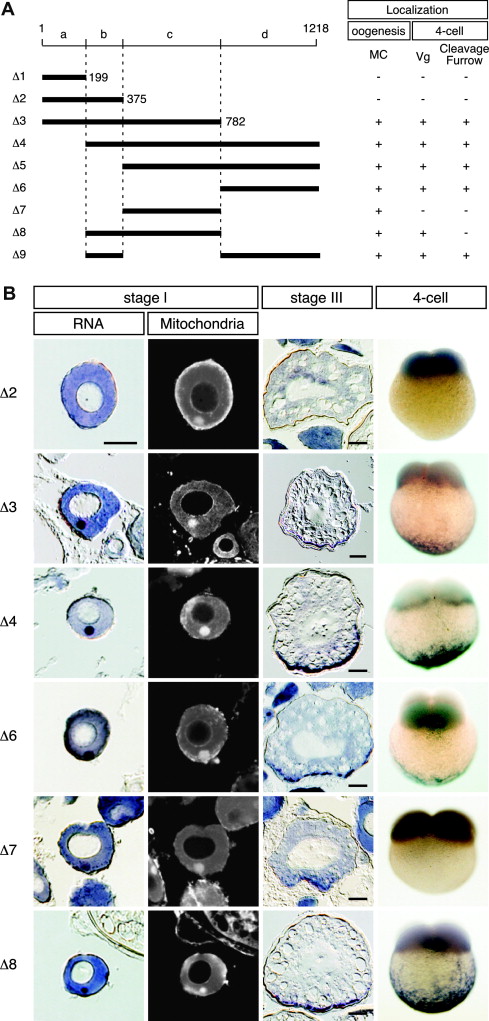
Fig. 6 Identification of cis-elements required for localization of dazl mRNA. (a) Schematic representation of deletions of zebrafish dazl 3′ UTR (Δ1–Δ9) used in transgenic experiments. Numbers represent nucleotide positions relative to the stop codon. Regions of the dazl 3′ UTR labeled a, b, c and d contain nucleotides 1-199, 200-375, 376-782, and 783-1218, respectively. The results of mRNA distribution analyses are indicated to the right of the schematic diagram. (+), normal pattern of distribution; (-), not localized. MC shows MC localization in oocytes. Vg shows localization at vegetal pole in 4-cell embryos. (b) Detection of the GFP mRNA fused with mutant 3′ UTRs in transgenic oocytes (stages I and III) and 4-cell embryos. For stage I oocytes, in situ hybridization was performed on sections counterstained with Mitotracker to detect mitochondria. Oocyte sections were cut at a thickness of 10 μm. Scale bars, 50 μm. A lateral view of the 4-cell embryo is shown.
Reprinted from Mechanisms of Development, 124(4), Kosaka, K., Kawakami, K., Sakamoto, H., and Inoue, K., Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis, 279-289, Copyright (2007) with permission from Elsevier. Full text @ Mech. Dev.