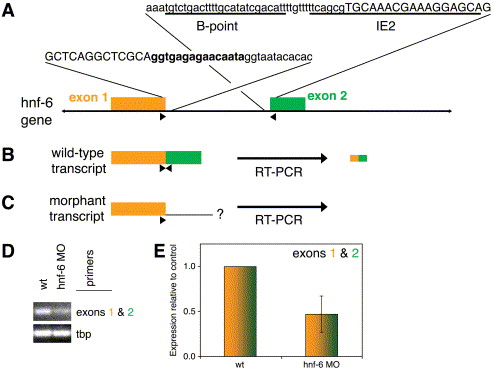
Fig. 5 Genomic organization and targeting of the zebrafish hnf-6 locus. (A) Genomic sequence and arrangement of zebrafish hnf-6. Two exons with adjacent intronic sequences are shown. Exon sequence is in CAPS, intron in lowercase. The sequence encoding the five amino acid insertion of the ‘β’ isoform is in bold. The sequences against which splice acceptor morpholinos were generated are underlined and labeled as IE2 and B-point, accordingly. Primer pair used for RT-PCR depicted in (B–D) is denoted by arrowheads. Diagram not to scale (intron approximately 4 kb). (B and C) Schematics of wild-type and morphant hnf-6 transcript, showing that the primer pair amplifies a fragment derived only from the wild-type transcript, as depicted in gold and green from the corresponding exons. (D and E) Semiquantitative (D) and quantitative (E) RT-PCR of cDNA from 24 hpf control (“wt”) and IE2-injected morphant embryos. There is a 50% reduction in the amplification of the cDNA fragments spanning the exons 1 and 2 splice site in morphant embryos compared with wild type. For all quantitative PCR experiments, hnf-6 cDNA amplification is standardized using amplification of the TATA-box binding protein (tbp) cDNA.
Reprinted from Developmental Biology, 274(2), Matthews, R.P., Lorent, K., Russo, P., and Pack, M., The zebrafish onecut gene hnf-6 functions in an evolutionarily conserved genetic pathway that regulates vertebrate biliary development, 245-259, Copyright (2004) with permission from Elsevier. Full text @ Dev. Biol.