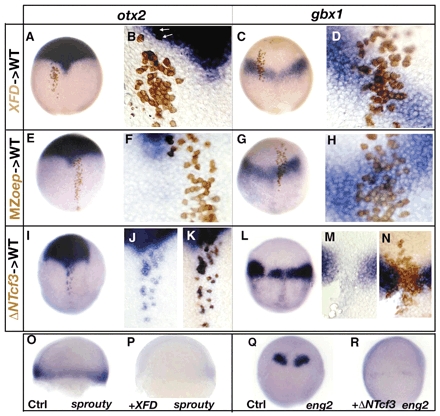
Fig. 3 Role of FGF, Nodal and Wnt signals in defining the otx2 and the gbx1-expression domain. (A-D) Embryo containing cells derived from injected embryos with a lineage tracer (brown) and XFD RNA (200 pg) stained for otx2 (A,B) or for gbx1 expression (C,D) at 90%. (B,D) Close-up of transplanted cells. XFD-transplanted cells (blind for FGF signaling) strictly follow the host otx2 expression domain or gbx1 expression domain. (E-H) Embryos containing cells derived from MZoep-injected embryos with a lineage tracer (brown) stained for otx2 (E,F) or for gbx1 expression (G,H) at 90%. (F,H) Close-up of transplanted cells. MZoep cells (blind for Nodal signaling) strictly follow the host otx2 expression domain or gbx1 expression domain. (I-N) Embryos containing cells derived from injected embryos with a lineage tracer (brown) and ΔNTcf3 RNA (400 pg). (I) Transplanted embryo stained for otx2. (J) Close-up of cells ectopically expressing otx2 followed by (K) staining the donor cells (brown). (L) Transplanted embryo stained for gbx1. (M) Close-up of the gap in the gbx1-expression domain followed by (N) staining the donor cells (brown). The ΔNTcf3-transplanted cells (blind for Wnt signaling) express otx2 ectopically and do not express gbx1. (O) Control embryo at 60% stained for sprouty4; (P) XFD-injected embryos do not express sprouty4, an FGF target gene. (Q) Control embryo at tailbud stained for eng2. (R) In ΔNTcf3-injected embryos, eng2 expression is strongly diminished. (A-R) Dorsal views, anterior upwards.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development