- Title
-
Hyperactive mTORC1 disrupts habenula function and light preference in zebrafish model of Tuberous sclerosis complex
- Authors
- Doszyn, O., Kedra, M., Zmorzynska, J.
- Source
- Full text @ iScience
|
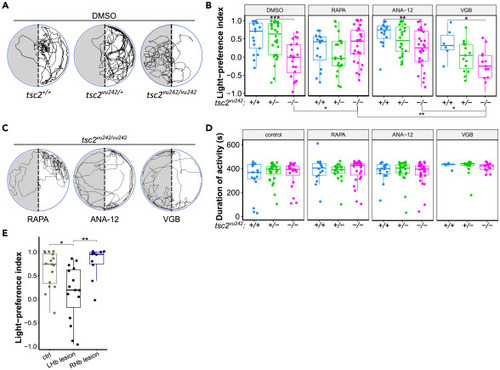
Light-preference test in tsc2vu242 fish (A) Exemplary tracks of tsc2vu242 fish from the light-preference test. (B) Light-preference index of tsc2vu242 fish after various treatments (tsc2vu242/vu242 vs. tsc2+/+: p = 0.0009 for DMSO, p = 0.588 for rapamycin (RAPA), p = 0.005 for ANA-12, p = 0.044 for vigabatrin (VGB); for tsc2vu242/vu242 treated with DMSO vs. RAPA p = 0.048). The dots on the boxplots represent the number of fish in the experiment (N > 10 per experimental group, except VGB tsc2+/+ where N = 6). (C) Exemplary tracks of tsc2vu242/vu242 mutant treated with RAPA, ANA-12, or VGB from the light-preference test. (D) Cumulative activity of tsc2vu242 fish during the light-preference test. The dots on the boxplots represent the number of fish in the experiment (N > 10 per experimental group, except VGB tsc2+/+ where N = 6). (E) Light-preference index of WT fish without lesions and with lesion of left (LHb) or right (Rhb) habenula (unlesioned vs. lesion of the left habenula: p = 0.019, lesion of the left habenula vs. lesion of the right habenula: p = 0.009). The dots on the boxplots represent the number of fish in the experiment (N > 10 per experimental group). |
|
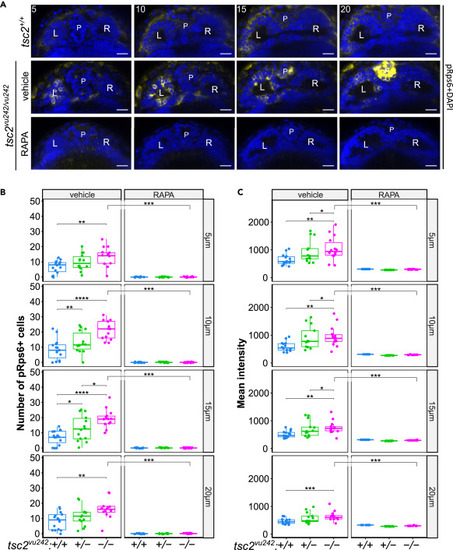
mTorC1 activation in tsc2vu242 fish habenulas (A) Representative optical sections through habenula of tsc2vu242/vu242 fish and their wild-type tsc2+/+ siblings at 5, 10, 15, and 20 μm from the top. pRps6—yellow, nuclei—blue. L—left habenula, R—right habenula, P—pineal complex. Scale bars, 20 μm. (B) Number of pRps6-positive cells in LdHb of tsc2vu242 fish (5 μm: tsc2vu242/vu242 control vs. tsc2+/+ control p = 0.004 and vs. tsc2vu242/vu242 treated with RAPA p = 1.038 × 10−5; 10 μm: tsc2vu242/vu242 control vs. tsc2+/+ control p = 4.656 × 10−5 and vs. tsc2vu242/vu242 treated with RAPA p = 1.038 × 10−5; 15 μm: tsc2vu242/vu242 control vs. tsc2+/+ control p = 1.584 × 10−5 and vs. tsc2vu242/vu242 treated with RAPA p = 1.038 × 10−5; 20 μm: tsc2vu242/vu242 control vs. tsc2+/+ control p = 0.004 and vs. tsc2vu242/vu242 treated with RAPA p = 1.038 × 10−5). The dots on the boxplots represent the number of fish in the experiment (N > 10 per experimental group). (C) Quantification of mean intensity of pRps6 fluorescence from LdHb of tsc2vu242 fish (5 μm: tsc2vu242/vu242 control vs. tsc2+/+ control p = 0.0051 and vs. tsc2vu242/vu242 treated with RAPA p = 1.0912 × 10−5; 10 μm: tsc2vu242/vu242 control vs. tsc2+/+ control p = 0.0017 and vs. tsc2vu242/vu242 treated with RAPA p = 1.0912 × 10−5; 15 μm: tsc2vu242/vu242 control vs. tsc2+/+ control p = 0.001 and vs. tsc2vu242/vu242 treated with RAPA p = 1.0912 × 10−5; 20 μm: tsc2vu242/vu242 control vs. tsc2+/+ control p = 0.0007 and vs. tsc2vu242/vu242 treated with RAPA p = 1.0912 × 10−5). The dots on the boxplots represent the number of fish in the experiment (N > 10 per experimental group). |
|
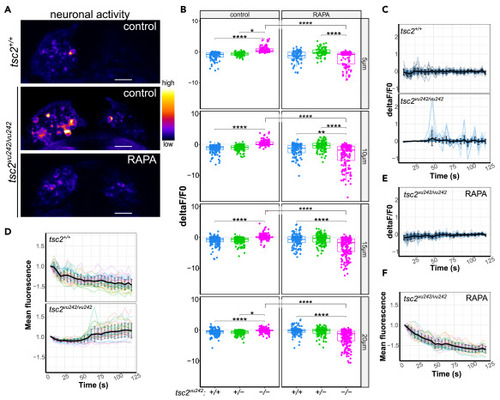
Neuronal activity in LdHb of tsc2vu242 fish (A) Representative images of neuronal activity in the habenulae of tsc2vu242 fish at 10 μm from the top. Scale bars, 20 μm. (B) Cumulative activity of the tsc2vu242 LdHb for all the layers of LdHb (5 μm: tsc2vu242/vu242 control vs. tsc2+/+ control p = 6.528 × 10−33 and vs. tsc2vu242/vu242 treated with RAPA p = 5.958 × 10−34; 10 μm: tsc2vu242/vu242 control vs. tsc2+/+ control p = 1.1232 × 10−44 and vs. tsc2vu242/vu242 treated with RAPA p = 1.625 × 10−47; 15 μm: tsc2vu242/vu242 control vs. tsc2+/+ control p = 5.928 × 10−31 and vs. tsc2vu242/vu242 treated with RAPA p = 4.68 × 10−52; 20 μm: tsc2vu242/vu242 control vs. tsc2+/+ control p = 1.104 × 10−14 and vs. tsc2vu242/vu242 treated with RAPA p = 5.958 × 10−34). Dots on boxplots represent single cells. (C) Neuronal activity change over time in the tsc2vu242 LdHb at 10 μm from the top. (D) Normalized mean GCaMP5F fluorescence over time in the tsc2vu242 LdHb at 10 μm from the top. (E) Neuronal activity change over time in the tsc2vu242/vu242 mutant’s LdHb at 10 μm from the top after RAPA pretreatment. (F) Normalized mean GCaMP5F fluorescence over time in the tsc2vu242/vu242 mutant’s LdHb at 10 μm from the top after RAPA. (C-F) The mean with SD is shown in black. 7 fish per genotype per treatment were analyzed (N = 7 per experimental group). |
|
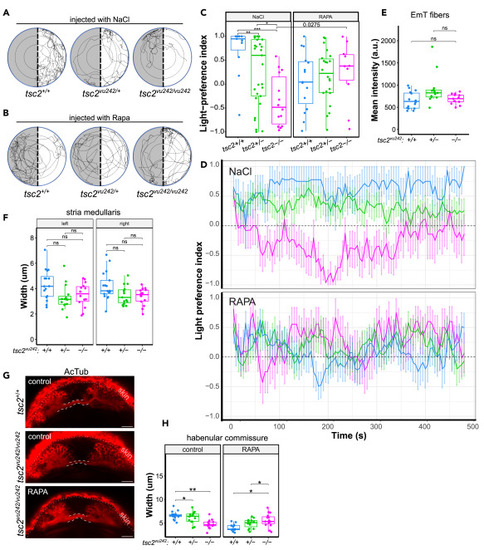
Rapamacin injections to the left habenula and afferent connectivity of LdHb in tsc2vu242 fish (A) Exemplary tracks of the tsc2vu242 fish injected to the left habenula with NaCl from the light-preference test. (B) Exemplary tracks of the tsc2vu242 fish injected to the left habenula with 1 μM rapamycin (Rapa) from the light-preference test. (C) Light-preference index of the tsc2vu242 fish after injections to the left habenula (tsc2vu242/vu242 vs. tsc2+/+: p = 0.00015 and vs. tsc2+/−: p = 0.006 for NaCl, p = ns for rapamycin (RAPA); tsc2vu242/vu242 fish injected with RAPA vs. NaCl: p = 0.0275). The dots on the boxplots represent the number of fish in the experiment (N > 10 per experimental group). (D) Analysis of light-preference index over time of the tsc2vu242 fish after injections to the left habenula (data presented as mean ± SD). (E) Mean intensity of calretinin fluorescence in the left habenulae of the tsc2vu242 fish. The dots on the boxplots represent the number of fish in the experiment (N > 10 per experimental group). (F) Width of SM in the tsc2vu242 fish brains. The dots on the boxplots represent the number of fish in the experiment (N > 10 per experimental group). (G) Representative horizontal optical sections through HC (outlined) of the tsc2vu242 fish (projection of 8 z-stacks). Scale bars, 20 μm. (H) HC width of the tsc2vu242 fish (tsc2vu242/vu242 vs. tsc2+/+: p = 0.002 for DMSO, p = 0.011 for RAPA). The dots on the boxplots represent the number of fish in the experiment (N > 10 per experimental group). |