- Title
-
Anterior movement of ventral diencephalic precursors separates the primordial eye field in the neural plate and requires cyclops
- Authors
- Varga, Z.M., Wegner, J., and Westerfield, M.
- Source
- Full text @ Development
|
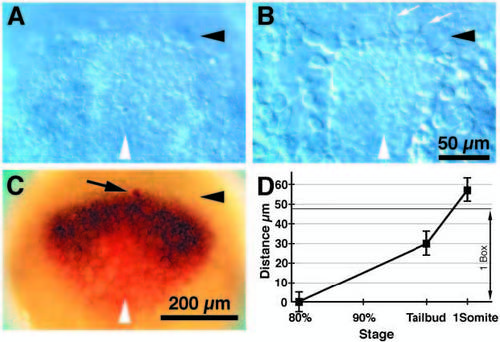
The anterior borders of the neural plate, the prechordal plate and the opl expression domain colocalize at 80% epiboly. (A-C) Dorsal views, anterior to the top, 80% epiboly, white arrowheads indicate midline; (A-B), Nomarski optics. (A) Dorsal view of live embryo focused at the level of the ectoderm. The anterior border of the neural plate is apparent as a thickening of the ectoderm (black arrowhead). (B) A deeper focal plane view of the embryo pictured in A. Cells with filopodia (white arrows) are apparently migrating at the anterior edge (black arrowhead) of the axial mesendoderm. (C) An ectodermal cell was labeled with lineage tracer (arrow, red cell) superficial to the anterior edge of the migrating axial mesendoderm (black arrowhead), and the embryo was immediately fixed and hybridized with RNA probes for opl (black expression domain) and otx2 (red expression domain). This demonstrated that at 80% epiboly, the anterior edges of the axial mesendoderm, the neural plate, and the opl expression domain coincide. (D) Distance between the anterior edge of the axial mesendoderm and the anterior edge of the neural plate between the 80% epiboly and 1-somite stages. The distance between these two morphological landmarks can be used for precise staging of the embryos as the axial mesendoderm advances relative to the anterior edge of the neural plate and the tailbud. 80% epiboly, n=9; tailbud stage, n=14; 1-somite stage, n=10. Distance = average ± s.d. in μm. 1 Box, the length of a box in the 10x10 reticule used for the fate map (see also Fig. 2B). Time scale is approximately linear with developmental time. Scale bars, 50 μm (A-B), 200 μm (C). EXPRESSION / LABELING:
|
|
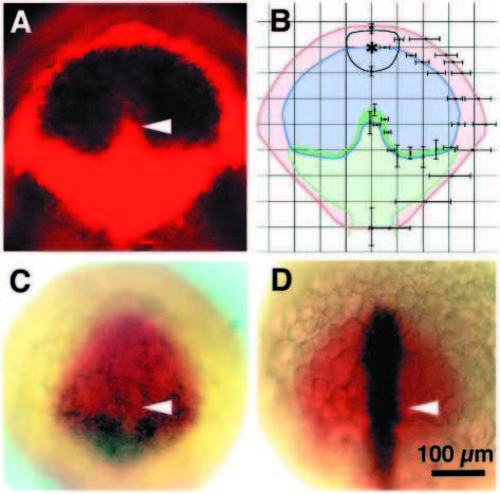
Gene expression domains subdivide the anterior neural plate. Dorsal views anterior to the top; (A,B,D) tailbud stage, (C) 1-somite stage; (A) The opl (black) expression domain is contained within the otx2 (red) domain as shown by fluorescence microscopy. The white arrowhead points to the indentation in the opl expression domain. (B) 10-15 embryos were doubly labeled with mRNA probes for otx2 and opl or mar. Gene expression borders were recorded on 10x10 grids representing the reticule in the eyepiece of the microscope. The distances from the midline and the origin (⋆) were measured in individual embryos and average values and standard deviations calculated. Such analysis produced averaged gene expression domains for otx2 (red), opl (blue) and mar (green). The averaged outline of the polster is indicated in black. Error bars indicate standard deviations. (C) opl (red) and pax6 (black) occupy overlapping domains in the anterior neural plate. The posterior portion of the pax6 expression domain overlaps the lateral arms of the mar expression domain (not shown). Cells farther anterior express pax6 less intensely and also express opl. A white arrowhead indicates the region corresponding to the mar protrusion and the indentation in the posterior border of the opl expression domain. (D) Cells of the prechordal plate and median neural plate express the cyclops gene (black). The site of the mar protrusion (white arrowhead) is not distinguished by cyclops gene expression pattern. opl expression is shown in red. Scale bar, 100 μm. EXPRESSION / LABELING:
|
|
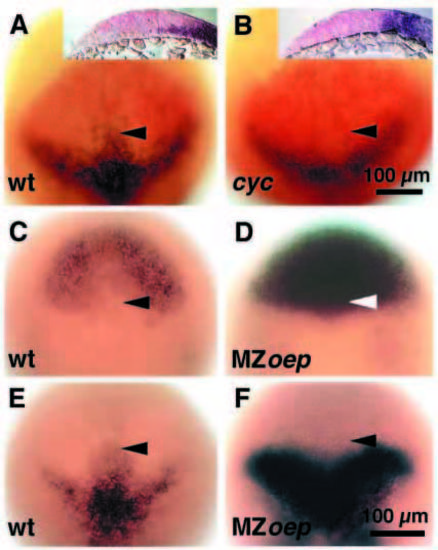
Opl and mar share a border of gene expression at tailbud stage that is perturbed in cyclops and MZoep mMutant embryos. Dorsal views; anterior to the top; insets in A and B, sagittal sections with dorsal to the top and anterior to the left. (A) Wild-type embryo labeled with mRNA probes for opl (red) and mar (black). The anterior protrusion of the mar expression domain fits into the indentation at the posterior border of the opl expression domain (arrowhead). The inset shows that the protrusion of mar-expressing cells is wedge-shaped beneath the posterior part of the opl domain. (B) cyclops mutant embryo labeled with mRNA probes for opl (red) and mar (black). The indentation in the opl expression domain and the protrusion in the mar domain are absent (arrowhead). Inset, no wedge shaped protrusion is visible; instead, a vertical border forms between the mar and opl expression domains. (C,E) Wild-type embryos labeled with mRNA probes for opl (C) and mar (E). The arrowheads point to the indentation at the posterior border of the opl expression domain (C) and to the anterior protrusion in the mar expression domain (E). (D,F) MZoep mutant embryos labeled with mRNA probes for opl (D) and mar (F). The indentation in the opl expression domain (D) and the protrusion in the mar domain (F) are absent (arrowheads). The dorsal axis is shortened in MZoep mutant embryos. Scale bar, 100 μm (A-F and insets). EXPRESSION / LABELING:
|
|
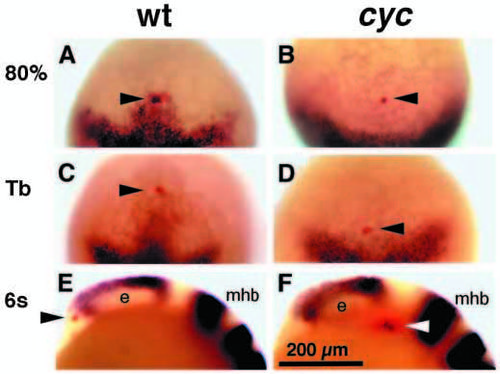
Wild-type cyclops gene function is required for anterior movement of ventral diencephalic precursors. (A-D) Dorsal views, anterior to the top. (E,F) Side views, anterior to the left, dorsal to the top. e, eye; mhb, mid-hindbrain boundary. (A,B) Single cells in corresponding regions of the neural plate of wild-type (A, arrowhead) and cyclops mutant embryos (B, arrowhead) were injected with rhodamine dextran. The first group of embryos (n=9 wild type; n=3 cyclops) was immediately fixed and labeled for mar expression by mRNA in situ hybridization. (C,D) The second group of embryos (n=7 wild type; 2 cyclops) was fixed 2 hours later and labeled for mar expression. (C) By this time the injected cell (arrowhead) had shifted anteriorly, and had stopped expressing mar in wild-type embryos. (D) In cyclops mutant embryos, the injected cell failed to move anteriorly and remained near the anterior border of the mar expression domain. (E,F) The third group (n=6 wild type; n=3 cyclops) was fixed 4 hours after injection. These embryos were labeled (black) to detect the expression of pax2 in the prospective optic stalk region, emx1 in the prospective telencephalon, eng3 in the prospective midbrain hindbrain border and krx20 in the prospective hindbrain. In wild-type embryos (E), the injected cell (red, arrowhead) was found anterior and ventral to the eye primordia. The ventral diencephalon had formed. (F) In cyclops mutants, the injected cell failed to move forward and remained posterior to the eye (arrowhead). cyclops mutants lacked ventral diencephalon. Scale bar, 200 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
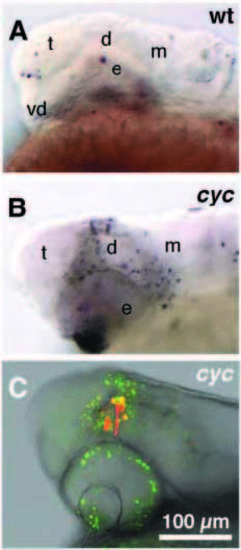
Cells that fail to migrate in cyclops mutant embryos die. (A,B) bright field; (C) superimposed confocal and bright-field image; side views, anterior to left, dorsal to the top, 24h. (A) TUNEL labeling of wildtype embryo. Cell death is observed only in the surface ectoderm and the lens (out of focus). No cell death is apparent in forebrain or midbrain. Similar results were obtained in 19 other embryos. (B) TUNEL labeling reveals dying cells dorsal and posterior to the eye of cyclops mutant embryos (n=7). (C) Embryos (n=5) doubly labeled with lineage tracer (rhodamine dextran, red) and Acridine Orange (AO, green) showed that diencephalic precursors failed to move forward to separate the single retinal field. Lineage tracer injected cells remained posterior / dorsal to the eyes and died in dorsal diencephalic and pretectal regions (yellow, doubly labeled with AO and lineage tracer). A few hours later all rhodamine labeled cells were doubly labeled with AO (not shown). In wild-type embryos, lineage tracer labeled cells were unlabeled by AO (n=12). Prospective brain regions: t, telencephalon; vd, ventral diencephalon; d, dorsal/posterior diencephalon; e, eye; m, midbrain. Scale bar, 100 μm. |
|
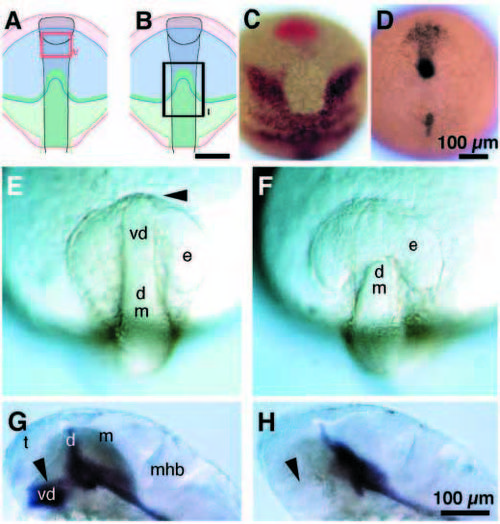
Diencephalic precursors are required for separation of the single eye field. (A-F) Dorsal views, anterior to the top; (A-D) tailbud stage. (A,B) Diagram showing gene expression domains: otx2, red; opl, blue; mar, green; cyclops, grey. (A) The Type V ablation is marked by the red rectangle. Type V ablations, like unoperated and sham ablations never produced cyclopia. (B) The Type I ablation at neural plate stage is marked by the black rectangle. This type of ablation removed the entire mar protrusion and flanking regions. (C) Embryo with Type I ablation, fixed immediately after operation and hybridized in situ with probes for hgg-1 (red, polster; Thisse et al., 1994) and mar (black) mRNAs. (D) Embryo with Type I ablation, fixed immediately after operation and hybridized in situ with a probe for cyclops mRNA. (E,F) 12-somite stage. (E) Unoperated embryo. Retinal precursors had separated in unoperated embryos by this stage (n=15), and only the optic stalk (arrowhead) connected the eye primordia. Sham operated embryos (n=9) are indistinguishable from unoperated siblings (not shown). (F) Cells in the protrusion of the mar expression domain were ablated at neural plate stage (Type I as in A-C). A single retinal field persisted in operated embryos (12-somite stage, n=15), which closely resembled the morphology of cyclops mutant embryos at the same developmental stage (not shown). (G,H) 30h, side views, anterior to the left, dorsal to the top. (G) shh expression marks ventral diencephalon in sham (not shown) and unoperated embryos (arrowhead). (H) Operated embryos (Type I ablation; 10 out of 13 embryos) lack a ventral diencephalon, as indicated by the absence of sonic hedgehog (black) expressing cells (arrowhead) at 30h, and are completely cyclopic. Prospective brain regions: t, telencephalon; e, primordial eye (field); vd, ventral diencephalon; d, dorsal/posterior diencephalon; m, midbrain; mhb, mid-hindbrain boundary. Scale bars, 100 μm. |