- Title
-
Harmonizing model organism data in the Alliance of Genome Resources
- Authors
- Alliance of Genome Resources Consortium
- Source
- Full text @ Genetics
|
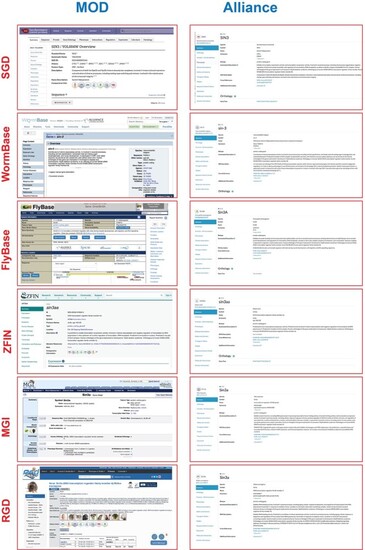
The Alliance Portal provides a harmonized view of research organism information. Left, current MOD pages; Right, current Alliance release 4.0 gene pages. |
|
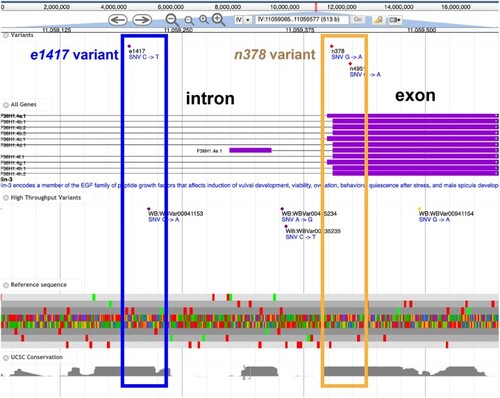
Example of Alliance JBrowse. The top is the standard control bar. Next are curated variants, often with known phenotypic consequences. The gene structure models (introns and exons) for each gene are shown, with the high throughput variants shown in the next track. The reference sequence in all reading frames is followed by a conservation track from University of California, Santa Cruz. Two alleles are highlighted in this figure: the blue box shows the |
|
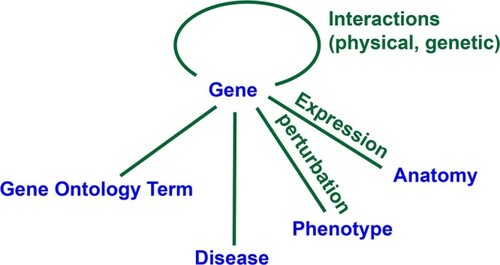
Conceptual map of gene-centered information. Perturbations of gene activity include alleles, variants, RNAi, knockdown, and transgenic overexpression. |
|
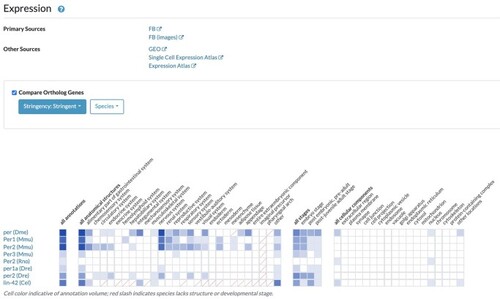
The expression widget. This example is for the |
|
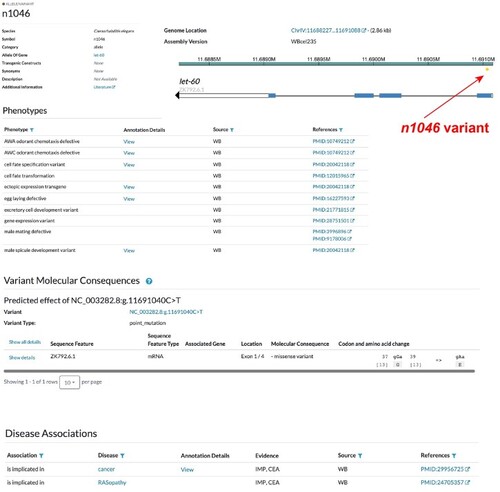
Montage of types of variant information and displays. The variant page has a summary, snapshot of Genomic Location, and then tables of Phenotypes, Molecular Consequences, and Disease Associations. |
|
Automatically generated gene summaries from structured data. Example of a gene summary for |
|
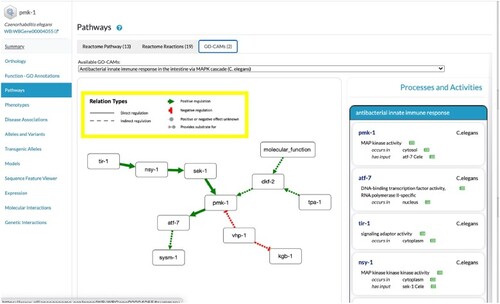
Views of pathways. GO-CAM model with simplified view for |
|
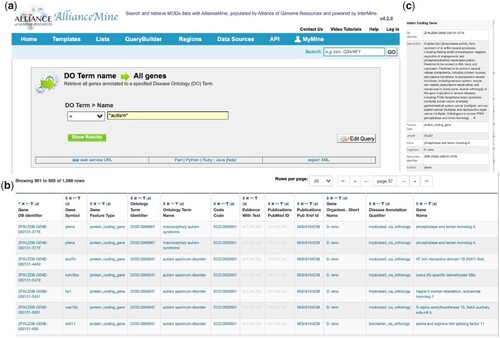
AllianceMine. Screen shots of AllianceMine output. Using a template query of disease ontology (DO) to all genes with the term “autism” a) returns 1088 genes b). Mousing over |