- Title
-
Hnrnpul1 controls transcription, splicing, and modulates skeletal and limb development in vivo
- Authors
- Blackwell, D.L., Fraser, S.D., Caluseriu, O., Vivori, C., Tyndall, A.V., Lamont, R.E., Parboosingh, J.S., Innes, A.M., Bernier, F.P., Childs, S.J.
- Source
- Full text @ G3 (Bethesda)
|
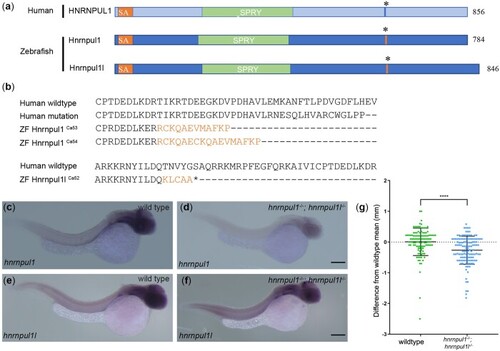
Mutation of human HNRNPUL1 and zebrafish EXPRESSION / LABELING:
PHENOTYPE:
|
|
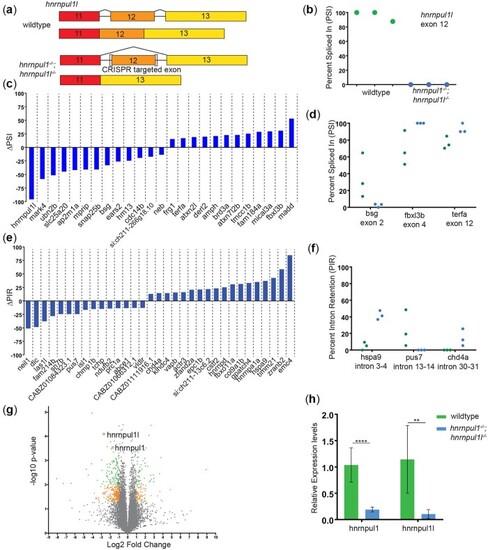
Loss of EXPRESSION / LABELING:
PHENOTYPE:
|
|
Loss of PHENOTYPE:
|
|
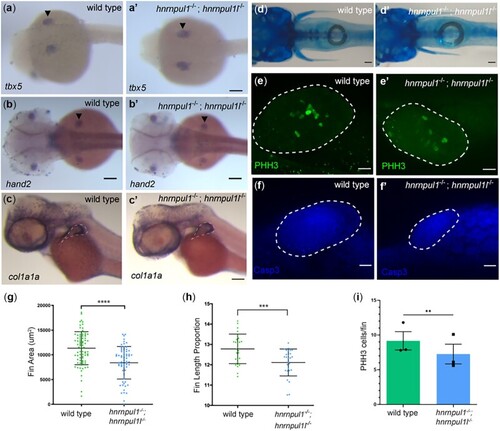
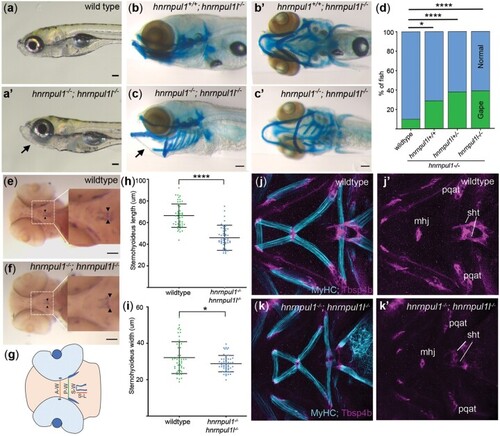
Musculoskeletal defects in EXPRESSION / LABELING:
PHENOTYPE:
|
|
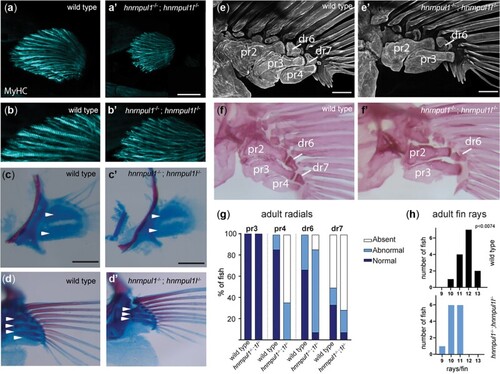
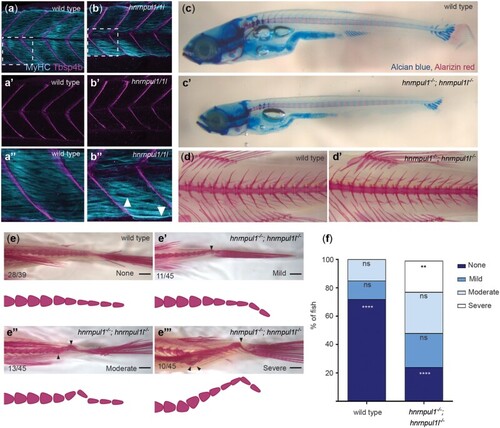
Loss of hnrnpul1l and hnrnpul1 leads to caudal scoliosis in adult Zebrafish. a, b) Muscle (MyHC, blue) and tendon (Tbsp4b, magenta) staining of 72 hpf wild type (a, a’’) and EXPRESSION / LABELING:
PHENOTYPE:
|
|
EXPRESSION / LABELING:
PHENOTYPE:
|
|
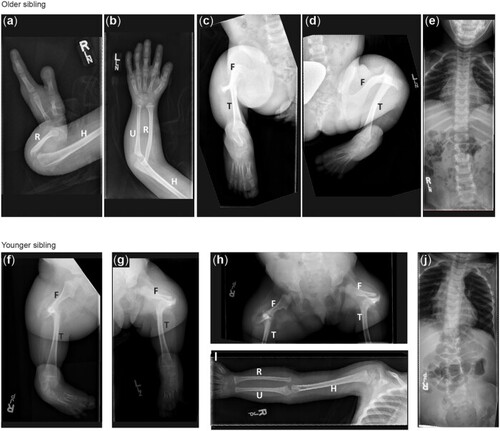
Radiographic features of siblings with a VUS in |

Unillustrated author statements PHENOTYPE:
|