- Title
-
Sumoylation regulates the assembly and activity of the SMN complex
- Authors
- Riboldi, G.M., Faravelli, I., Kuwajima, T., Delestrée, N., Dermentzaki, G., De Planell-Saguer, M., Rinchetti, P., Hao, L.T., Beattie, C.C., Corti, S., Przedborski, S., Mentis, G.Z., Lotti, F.
- Source
- Full text @ Nat. Commun.
|
|
|
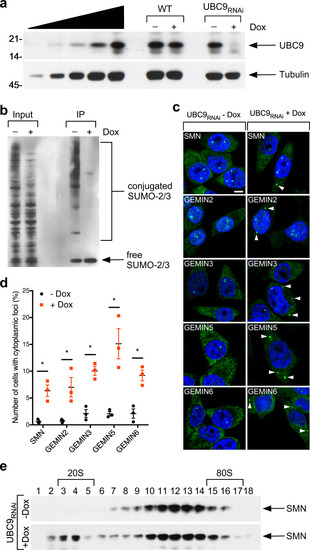
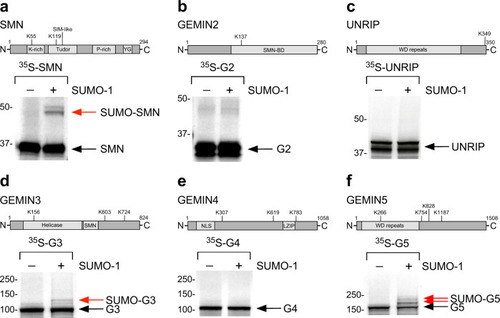
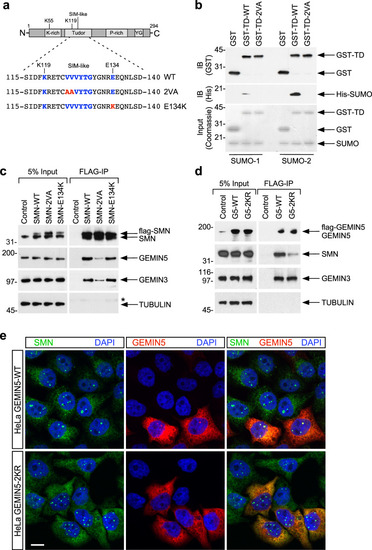
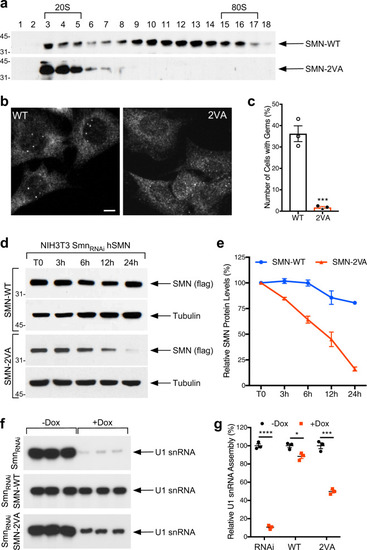
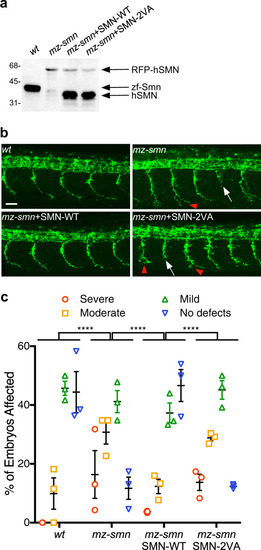
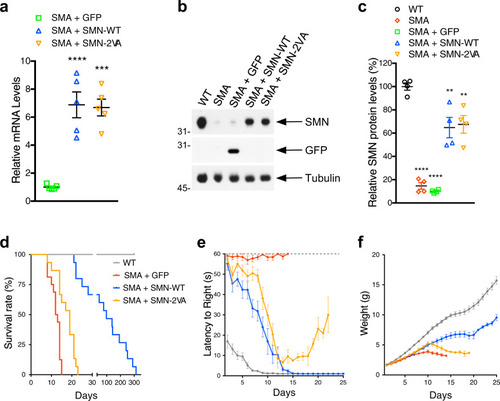
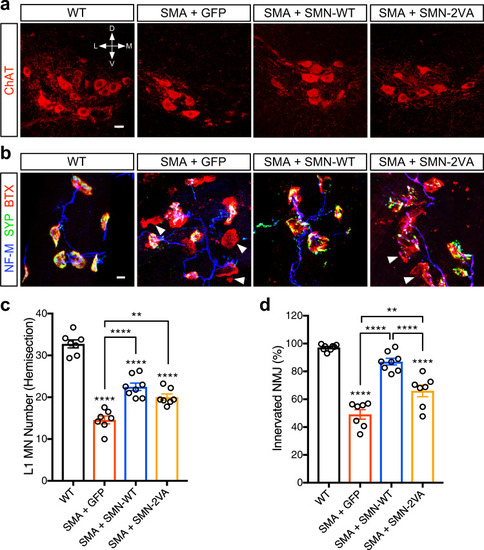
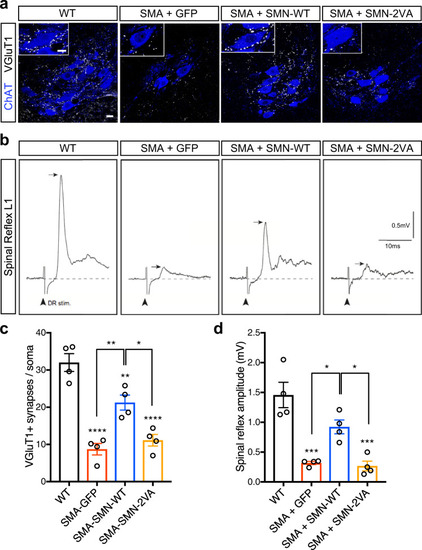
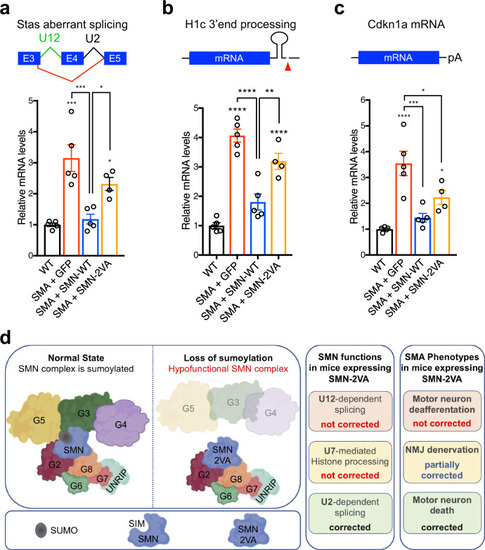
Schematic representation of the bioinformatically predicted sumoylation sites in SMN ( |
|
|
|
|
|
|
|
|
|
|
|
|
|
|