- Title
-
Lysosomal cholesterol accumulation contributes to the movement phenotypes associated with NUS1 haploinsufficiency
- Authors
- Yu, S.H., Wang, T., Wiggins, K., Louie, R.J., Merino, E.F., Skinner, C., Cassera, M.B., Meagher, K., Goldberg, P., Rismanchi, N., Chen, D., Lyons, M.J., Flanagan-Steet, H., Steet, R.
- Source
- Full text @ Genet. Med.
|
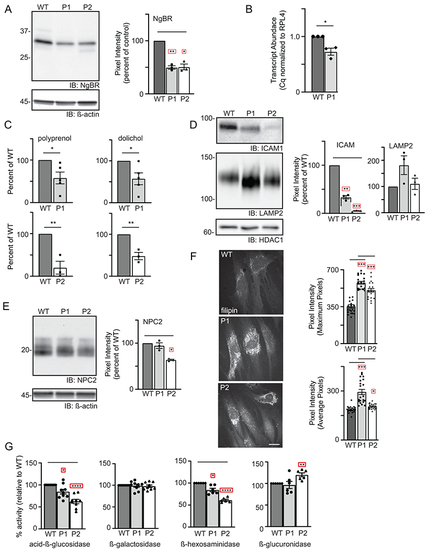
(A) Representative Western blot for NgBR in WT and patient fibroblasts and quantitation of NgBR levels relative to ß-actin (n=3). Error=S.E.M. Statistical analysis was performed using a Dunnett’s test. (B) Quantitative PCR analysis of NUS1 transcripts in WT and P1 fibroblasts (n=3). The number of WT vs. variant alleles is shown. (C) Analysis of total dolichol and polyprenol levels in WT and patient fibroblasts. Analysis of the two patient lines (P1; n=5 and P2; n=3) were performed at different times using separate WT cells. Statistical analysis was performed using an unpaired Student’s t test. (D) Representative Western blots for ICAM1 and LAMP2 in WT and patient fibroblasts and quantitation of protein levels relative to HDAC1 (n=3). Error=S.E.M. Statistical analysis was performed using a Dunnett’s test. (E) Representative Western blot for NPC2 in WT and patient fibroblasts and quantitation of levels relative to ß-actin (n=3). Error=S.E.M. Statistical analysis was performed using a Dunnett’s test. (F) Representative images of filipin-stained WT and patient fibroblasts and quantitation of pixel intensity in at least 40 different regions across 20 different cells. Error=S.E.M. Statistical analysis was performed using a Dunnett’s test. Scale bar = 10μm. (G) Protein-normalized activity of acid-ß-glucosidase, ß-hexosaminidase, ß-glucuronidase and ß-galactosidase in WT and patient fibroblasts. Arbitrary fluorescence units are plotted. Statistical analysis was performed using a Dunnett’s test. For all statistics *p<0.05, ** p<0.01, ***p<0.001. Red boxes indicate the additional correction (Dunnett’s test) was applied for comparison to a single control group. |
|
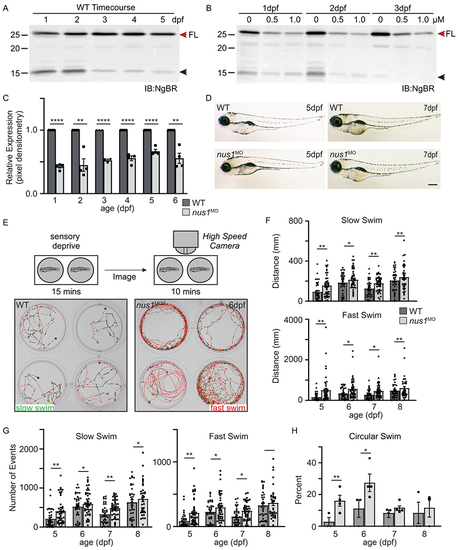
A) Western blots of 1-5dpf whole embryo lysates show NgBR is expressed during early development. Full length (FL) protein is indicated by a red arrowhead, as is a smaller processed piece. n=4 experimental replicates, 20 embryos per biological sample per experiment. B,C) Western blot show injection of 0.5 and 1.0 nM nus1 morpholino reduces NgBR abundance ~50% in embryos 1-6dpf. n=4 experimental replicates, 20 embryos per biological sample per experiment. D) Bright field images of WT and nus1 morphant (MO) embryos 5 and 7 dpf show no outward physical defects. n=100 embryos analyzed from 3 experiments. Scale bar=100μm. E) Schematic describes workflow for Zebrabox analyses of embryo motility. Image of 4 representative wells are shown from one plate containing WT and nus1 morphants. Traced swim patterns indicate the total path swam by the embryo, as well as swim speed. Red lines indicate a high velocity (fast) swim, while green lines indicate lower velocity (slow) swim. Images show nus1 morphants spend more time swimming at higher velocity than WT, often swimming in a circle around the well. F) Graphs summarizing the distance swam by an individual embryo (one dot) while at the fast or slow speed from 5-8 dpf. G) Graphs illustrate the number of new swim events initiated by an individual fish (one dot) while at the slow or fast swim speed from 5-8 dpf. H) Graphs illustrate the percent of total embryos in 4-5 experiments that exclusively swim in a circle at the edge of the well. n=50-100 embryos. For all quantitation: n= 50-75 embryos over 4-6 independent experiments. Error= S.E.M., significance calculated by the student’s t-test where *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. |
|
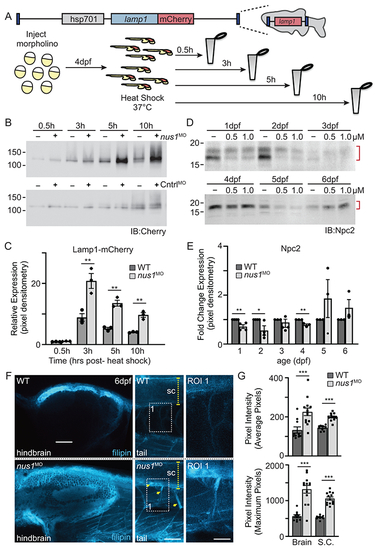
A) Schematic illustrates transgene in zebrafish genome and the workflow for analyzing Lamp1 in WT and nus1 morphant embryos. As shown, the transgene encodes a fusion between lamp1 and monomeric Cherry (mCherry); expression is controlled by the heat shock promoter (hsp701). B,C) Western blot analysis and graphic quantitation of Lamp1 over a time course (0.5-10h) following heat shock induction of its expression show while similar amounts of Lamp1-mCherry are made post-heat shock, protein accumulates in nus1 morphants. D,E) Western blot analysis and graphic quantitation of Npc2 abundance in WT and morphants (using either 0.5 or 1.0 μM morpholino) from 1-6dpf. Red bracket indicate Npc2 doublet; both bands included in the quantitation of relative abundance. For all graphs: n=3 experiments, with 25 embryo per sample per experiment. Error=S.E.M, significance calculated by the student’s t-test where *p<0.05, **p<0.01. F) Confocal analyses of filipin stained WT and nus1 morphant (MO) embryos 6dpf show cholesterol accumulation in the hindbrain, spinal cord (sc), and motor axons of morphant embryo tails. Yellow arrowheads highlight motor neuron cell bodies in ventral spinal cord and their associated axonal projections. Boxed region represents a region of interest (ROI) magnified in the right panel. Scale bars = 50 and 25 μM, respectively. G) Graph represents quantitation of the average and maximum pixel intensities from the hindbrains, spinal cords, and axons of 10-15 embryos from n=3 independent experiments. Error=S.E.M, significance calculated by the student’s t-test where ***p<0.001. |
|
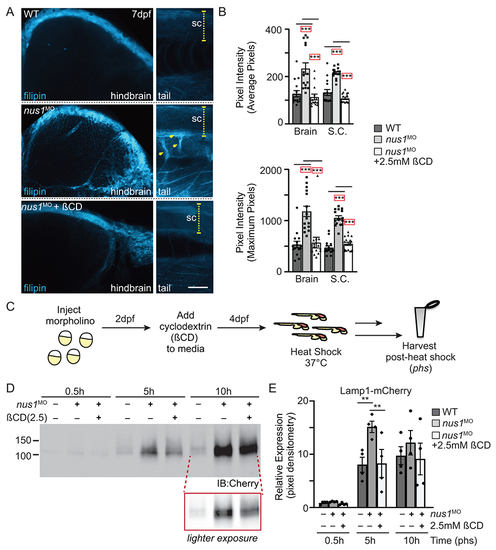
A) Confocal analyses of filipin stained WT, nus1 morphant (MO), and ßCD treated nus1 morphant embryos show reduced cholesterol accumulation in the hindbrain, spinal cord (sc), and motor axons of morphant embryos 7dpf. Yellow arrowheads highlight axonal projections motor neurons in ventral spinal cords of nus1 morphants, which are not detected with filipin staining in either WT or ßCD treated embryos. Scale bars =50μM. B) Graph represents quantitation of pixel the average and maximum intensities from hindbrain, spinal cords, and axons of 15 embryos from n=3 experiments. Error=S.E.M, significance calculated by the student’s t-test where ***p<0.001. C) Schematic illustrates workflow for analysis of Lamp1 expression following ßCD treatment. D,E) Western blot analysis and graphic quantitation of Lamp1 abundance (0.5-10h following heat shock induction of its expression) show Lamp1 accumulation is alleviated in nus1 morphants treated with 2.5 mM ßCD. n=4 independent experiments with 25 embryos per sample per experiment. Error=S.E.M, significance calculated by the student’s t-test where *p<0.05, **p<0.01. |
|
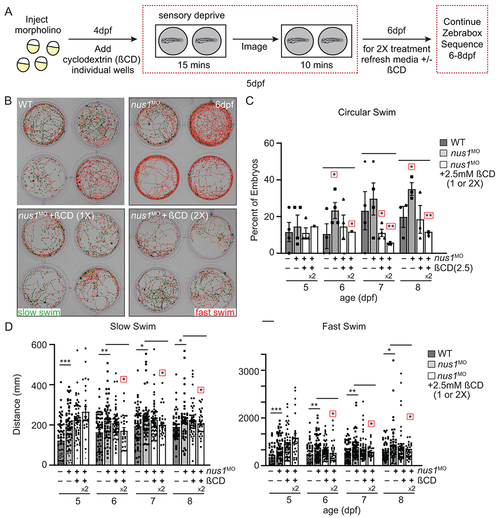
A) Schematic illustrates workflow for Zebrabox analyses of embryo motility following ßCD treatment. B) Images show 4 representative wells from 12 well plates containing WT, nus1 morphant, or nus1 morphant embryos treated 1 or 2x with 2.5mM ßCD. Traced swim patterns indicate the total path swam by the embryo, as well as swim speed. Red lines indicate a high velocity (fast) swim, while green lines indicate lower velocity (slow) swim. Images show treating nus1 morphants with ßCD reduces both their swim speed and the tendency to swim in a circle around the well. C) Graph showing percent of total embryos swimming exclusively in a circle. D) Graph showing distance swam by an individual embryo (black dot) at slow and fast swim speeds 5-8dpf. Data show that treating nus1 morphants with ßCD twice significantly reduces swim distance, restoring WT-like distances. For all graphs: n=3 experiments, with >25 embryos per “genotype” per condition. Error=S.E.M, significance calculated by the Dunnett’s test where correction for a single control sample was applied. *p<0.05, **p<0.01. ***p<0.0001. Red boxes indicate an additional correction (Dunnett’s test) was applied for comparison to a single control group. |