- Title
-
Trim46 contributes to the midbrain development via Sonic Hedgehog signaling pathway in zebrafish embryos
- Authors
- Jung, J., Kim, J., Huh, T.L., Rhee, M.
- Source
- Full text @ Animal Cells Syst (Seoul)
|
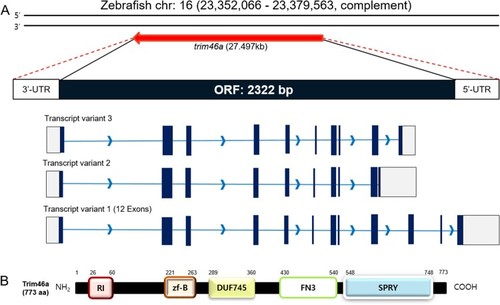
Schematic diagrams of the ORF of |
|
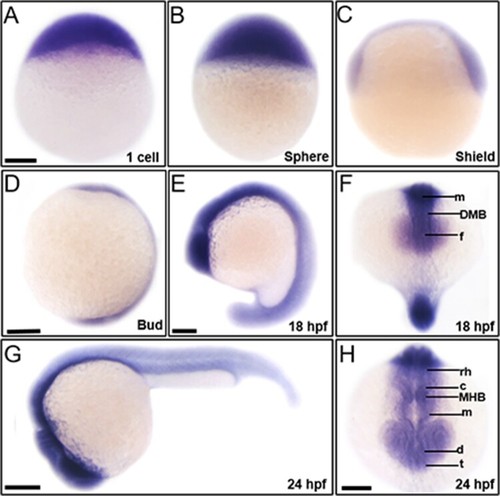
Spatiotemporal expression patterns of |
|
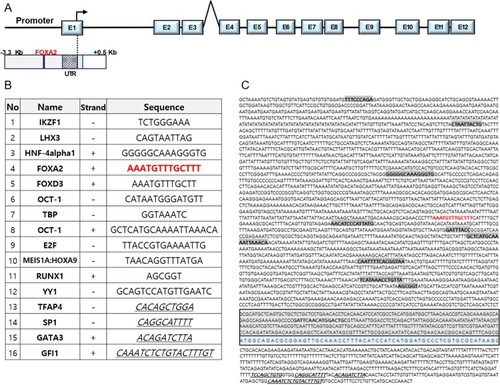
Putative |
|
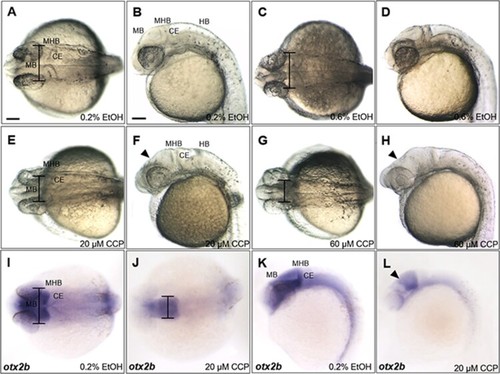
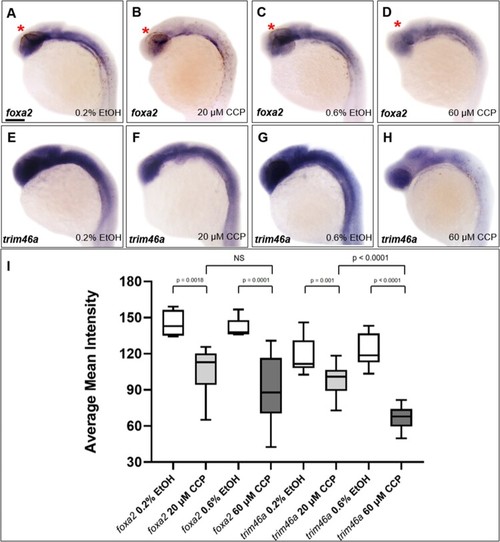
Inhibition of SHH signaling with cyclopamine resulted in developmental defects in the midbrain of zebrafish embryos at 24 hpf. (A–D) Zebrafish embryos at 24 hpf which were treated 0.2% and 0.6% EtOH from 4 hpf. 0.6% EtOH-exposed embryos showed similar phenotypes of midbrain and MHB in comparison to those of 0.2% EtOH-exposed embryos. (E and F) Cyclopamine-treated embryos (20 μM) showed reduction in width of the midbrain and dorsally shrunken midbrain in comparison to those of EtOH controls. (G and H) 60 μM cyclopamine-treated embryos exhibited severe defects in the midbrain. (I–L) Spatiotemporal expression patterns of otx2b in the midbrain of cyclopamine-treated embryos at 24 hpf. CCP, cyclopamine. (A–L) Scale bars: 50 μm. |
|
Inhibition of SHH signaling by cyclopamine decreased the level of |