- Title
-
Biodegradable Harmonophores for Targeted High-Resolution In Vivo Tumor Imaging
- Authors
- Sonay, A.Y., Kalyviotis, K., Yaganoglu, S., Unsal, A., Konantz, M., Teulon, C., Lieberwirth, I., Sieber, S., Jiang, S., Behzadi, S., Crespy, D., Landfester, K., Roke, S., Lengerke, C., Pantazis, P.
- Source
- Full text @ ACS Nano
|
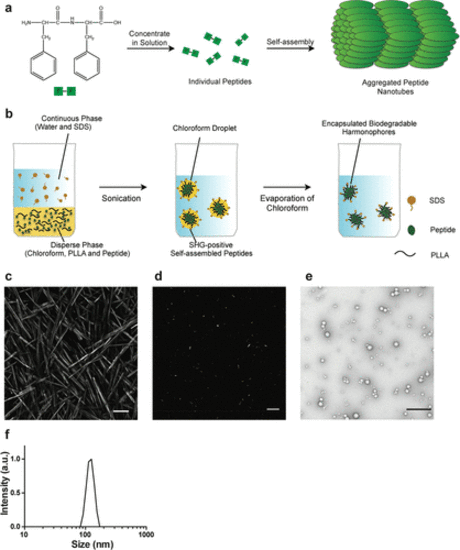
Synthesis and analysis of bioharmonophores. (a) Schematic of the self-assembling reaction of diphenylalanine peptides (FF) into large-scale nanotube structures from a concentrated solution. (b) Schematic of the emulsion–solvent evaporation method for the synthesis of bioharmonophores. Self-assembling peptides are dissolved in chloroform along with biodegradable poly(l-lactic acid) (PLLA) and emulsified with the surfactant sodium dodecyl sulfate (SDS) using sonication, followed by evaporation of chloroform. (c) SHG signal from diphenylalanine peptide nanotubes aggregated on top of the imaging chamber. Peptide nanotubes were illuminated with a 850 nm pulsed laser. Image composite of multiple stitched images. (d) SHG signal from encapsulated triphenylalanine peptide (FFF) bioharmonophores immobilized in 1% low melting agarose illuminated with 850 nm pulsed laser. (e) TEM image of synthesized FFF-based bioharmonophores showing uniform spherical nanoparticles. (f) DLS data showing the size distribution of synthesized bioharmonophores. Scale bar, 100 μm (c); 10 μm (d); 500 nm (e). |
|
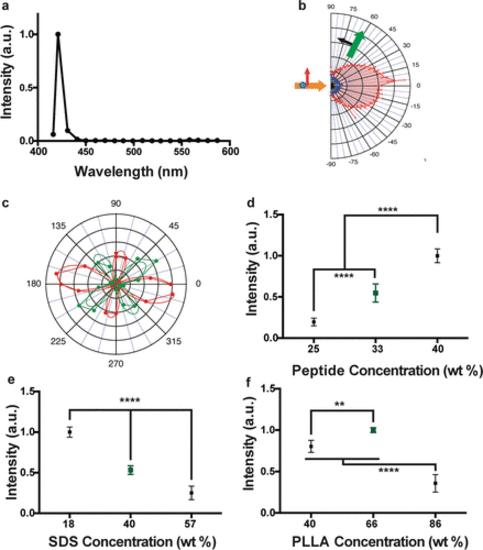
Optical characterization of bioharmonophores and analysis of parameters influencing bioharmonophore formation. (a) Normalized SHG signal spectrum of FFF-based bioharmonophores (signal ranging from 400 to 600 nm) illuminated with 850 nm pulsed laser. The characteristic SHG peak is centered around 425 nm. (b) SHG emission pattern of Triphenylalanine based bioharmonophores. Orange arrow indicates excitation beam direction. Green arrow shows SHG collection direction, which rotates between −90° and +90°. The detected polarization is in the beams plane (P, black arrow). The red pattern shows PPP polarization configuration (excitation and detection polarizations in the plane of the beams), and the blue pattern shows PSS (excitation with a perpendicular polarization). (c) SHG intensity vs incident polarization angle for a bioharmonophore, highlighted by the solid white circle in Supplementary Figure 5. Red color shows detection along the X axis while green color shows detection along the Y axis. The experimental curve is a dotted line, the corresponding fitted curve, assuming C2 symmetry, is a solid line. (d) Influence of using different amounts of FFF peptide during bioharmonophore production on the SHG signal intensity. The optimal condition (33 wt %) is marked in green. The use of higher FFF peptide amount leads to aggregates (n = 5). (e) Influence of SDS concentration (wt % of disperse phase) on SHG intensity of generated bioharmonophores. The optimal condition (40 wt % SDS) with high bioharmonophore stability and less aggregation is marked in green (n = 5). (f) Influence of using different amounts of PLLA during bioharmonophore production on the SHG intensity of the generated bioharmonophores. The optimal condition (66 wt % PLLA) is marked in green (n = 5). Mean ± s.d. ****, P < 0.0001, **, P < 0.005, *, P < 0.05 (ordinary one-way ANOVA with Tukey’s multiple comparisons). |
|
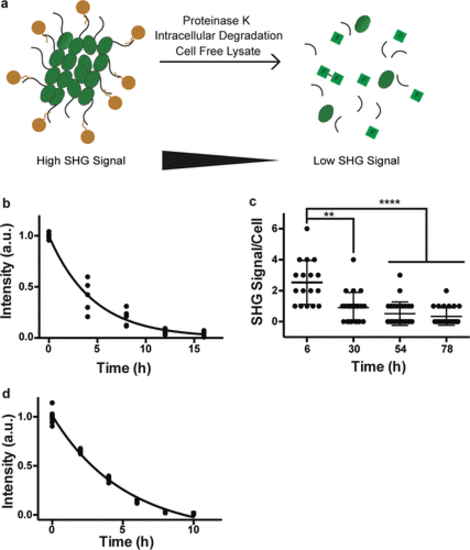
Bioharmonophores can be degraded by proteases, cells, and cell-free lysate systems. (a) Schematic showing different degradation methods utilized to assess biodegradability of the bioharmonophores. (b) Graph displaying the change of SHG signal intensity over time of bioharmonophores incubating with proteinase K (n = 5). Mean values of data points were fitted for one-phase exponential decay. (c) Quantification of SHG signal/cell after overnight incubation of Tat-peptide functionalized bioharmonophores over time. SHG signal/cell is significantly reduced 30 h after reseeding. Mean ± s.d. ****, P < 0.0001, **, P < 0.005, *, P < 0.05 (nonparametric Kruskal–Wallis test with Dunn’s post hoc multiple comparison). (d) Graph showing the loss of SHG signal intensity when bioharmonophores are subjected to the cell-free reticulate lysate degradation system (n = 5). Mean values of data points were fitted using a one phase exponential decay. Scale bar, 10 μm (c); 10 μm (d). |
|
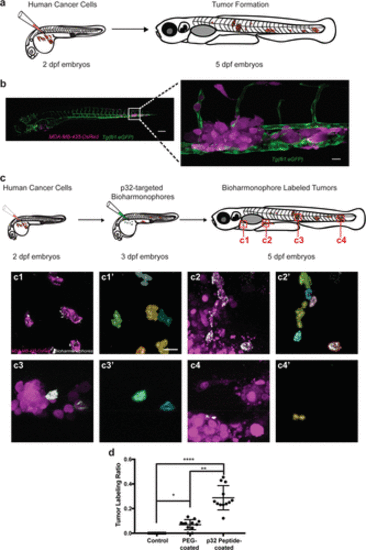
Bioharmonophores can be specifically targeted to single cancer cells in vivo. (a) Schematic showing the generation of a zebrafish cancer model by injecting MDA-MB-435-DsRed cancer cells into the Duct of Cuvier (DoC) at 2 dpf, resulting in tumors spread to multiple locations of the zebrafish body at 5 dpf. (b) Composite image of the cancer model (left) in a 5 dpf old zebrafish embryo. Close-up image of one of the tumor sites (right) reveals DsRed-labeled tumors (magenta), adjacent to the eGFP-labeled vasculature (green). (c) Schematic showing cancer cell injection of 2 dpf zebrafish embryos followed by bioharmonophore injection into DoC of 3 dpf zebrafish embryos and subsequent fluorescence and SHG imaging at 5 dpf. Red rectangles labeled as c1–4 denote the regions of interest that are illustrated in more detail. Individual panels showing the images of labeled cancer cells with the details of bioharmonophore (white) labeling down to single cancer cells (magenta) in solid tumors (c1–4). Colored cell boundary reconstruction of targeted cancer cells using the bioharmonophore SHG signal (c1′–4′). Note that cellular bioharmonophore distribution can in most cases predict cell morphologies. Scale bar, left panel 200 μm, right panel 20 μm (b); 15 μm (c). (d) Quantification of the fraction of SHG-labeled tumors as the ratio of labeled tumors to all tumors in a given zebrafish embryo after PEG- and p32 peptide-coated bioharmonophore injection, respectively. Each data point signifies one zebrafish. Note that active targeting with p32-coated bioharmonophores significantly increases the labeling efficiency (approximately 4-fold). Mean ± s.d. ****, P < 0.0001, **, P = 0.0063, *, P = 0.0470 (nonparametric Kruskal–Wallis test with Dunn’s post hoc multiple comparison). N = 12, pooled from 3 independent experiments. PHENOTYPE:
|