- Title
-
An ancestral 10-bp repeat expansion in VWA1 causes recessive hereditary motor neuropathy
- Authors
- Pagnamenta, A.T., Kaiyrzhanov, R., Zou, Y., Da'as, S.I., Maroofian, R., Donkervoort, S., Dominik, N., Lauffer, M., Ferla, M.P., Orioli, A., Giess, A., Tucci, A., Beetz, C., Sedghi, M., Ansari, B., Barresi, R., Basiri, K., Cortese, A., Elgar, G., Fernandez-Garcia, M.A., Yip, J., Foley, A.R., Gutowski, N., Jungbluth, H., Lassche, S., Lavin, T., Marcelis, C., Marks, P., Marini-Bettolo, C., Medne, L., Moslemi, A.R., Sarkozy, A., Reilly, M.M., Muntoni, F., Millan, F., Muraresku, C.C., Need, A.C., Nemeth, A.H., Neuhaus, S.B., Norwood, F., O'Donnell, M., O'Driscoll, M., Rankin, J., Yum, S.W., Zolkipli-Cunningham, Z., Brusius, I., Wunderlich, G., Genomics England Research Consortium , Karakaya, M., Wirth, B., Fakhro, K.A., Tajsharghi, H., Bönnemann, C.G., Taylor, J.C., Houlden, H.
- Source
- Full text @ Brain
|
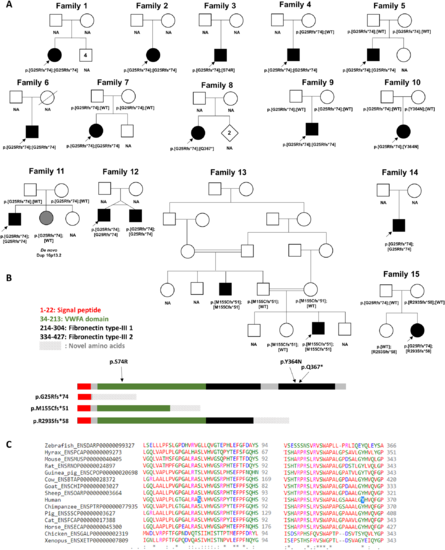
Pedigrees, variant localization and evolutionary conservation of missense changes in VWA1. (A) Pedigrees of the 15 families described here with predicted protein consequences of variants and the segregations pattern, where known. NA = DNA not available for testing. Filled symbols indicate early onset motor axonal neuropathy. Grey shading in Family 11 indicates a neurological presentation consistent with the observed duplication of 16p13.2. Genotype in younger twin in Family 12 was inferred because of monozygosity. (B) Schematic diagram showing the position of the variants identified in this study in relation to the protein domains in VWA1. The VWFA domain is shown in green whilst the two fibronectin type-III domains are shown in black. Figure is based on coordinates as listed in entry Q6PCB0 of the UniProt database (www.uniprot.org). (C) Evolutionary conservation of amino-acids in VWA1 orthologues at p.Ser74 and p.Asn364, the two sites where missense changes were identified. |
|
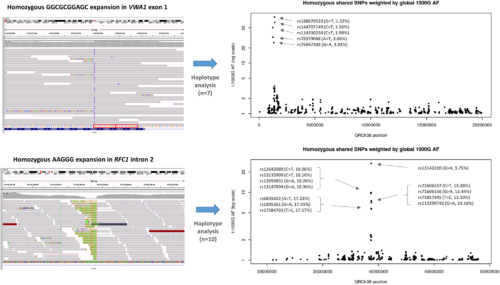
Haplotype analysis performed on individuals with homozygous repeat expansions in VWA1 (top) compared to similar results for RFC1 (bottom). Shared homozygous SNPs are plotted for 20 Mb segments of chromosome 1 and 4. In both cases, the most significant regions detected span the VWA1 and RFC1 loci where several consecutive SNPs are homozygous in 7/7 or 10/10 individuals respectively. For the VWA1 locus, the five shared SNPs labelled with rsIDs are highly informative, with global 1000 Genomes project allele frequencies <5% (1000G AF). The reciprocal of the allele frequency is plotted on the y-axis to give an idea of the information content for each of the shared SNPs. Within the VWA1 haplotype block we also observed a rare SNV in the 5′-UTR of ANKRD65 (rs758603246), which is heterozygous in 3/7 individuals. The variants appear to be in complete linkage disequilibrium as in the 100KGP all individuals with rs758603246 also have p.G25Rfs*74. Our interpretation is that rs758603246 is a more recent mutation and so is observed only on a subset of p.G25Rfs*74-containing haplotypes. |
|
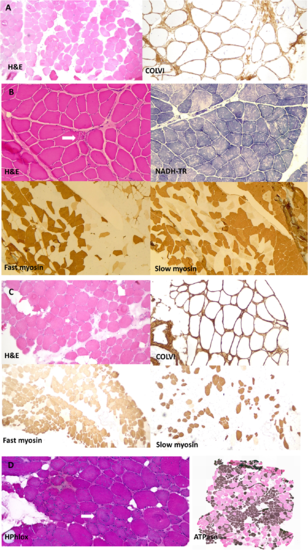
Muscle biopsy images from our patients all homozygous for p.G25Rfs*74. (A) Muscle biopsy (quadriceps) from a patient in Family 2 (performed at age 41 years). Images show marked fibre hypertrophy and increased fibrosis with endo and perimysial fatty infiltration. Labelling for collagen VI appears normal. (B) Muscle biopsy (right quadriceps) from a Family 4 patient (aged 6 years). Biopsy shows predominantly neuropathic aspects with neurogenic atrophy (arrow) and thickening of the endomysium and perimysium. Fascicular grouping of type 1 fibres implicates a chronic neurogenic process. NADH-TR shows the presence of moth-eaten fibres (non-specific finding). (C) Muscle biopsy (quadriceps) from a patient from Family 5 (aged 41 years). Marked variation in fibres size and increased endomysial fibrosis with mostly perimysial fatty infiltration. Rimmed vacuoles are shown with an arrow. There is a large predominance of type 2 fibres. Labelling for collagen VI appears normal. (D) Muscle biopsy from the index case in Family 12 (right vastus lateralis, age 52). Myopathic features are present, with marked fibre lobulation and variable degree of whorling of myofibrils (arrow). Endomysial fatty infiltration. Large areas of fibre grouping in this biopsy as well indicates a neurogenic aetiology. ATPase staining was performed at pH 4.2. H&E = haematoxylin and eosin. |
|
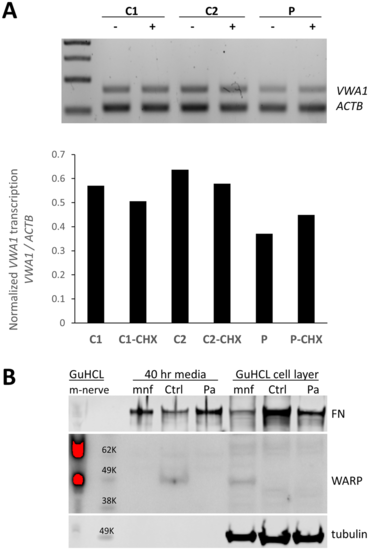
Consequences of the 10 bp insertion at the transcript and protein level. (A) Duplex PCR suggests a 10 bp insertion results in partial nonsense-mediated decay. RT-PCR products using primers for VWA1 and ACTB using RNA from proband in Family 11 (P) compared to control fibroblasts (C2, C2). The ratio of VWA1 to ACTB was reduced in the patient but was restored when patient fibroblasts were grown in presence of the nonsense-mediated decay inhibitor cycloheximide (CHX). (B) Immunoblotting indicated detectable levels of VWA1 (WARP) in human healthy control dermal fibroblasts; although the secreted VWA1 was not able to incorporated into ECM (cell layer), it was detectable in the conditioned medium. In contrast, no detectable VWA1 was seen in the patient’s dermal fibroblast in either the conditioned medium or the cell layer. A high level of VWA1 was detected in mouse sciatic nerve extraction (m-nerve). In the primary culture of mouse sciatic nerve (mnf), the VWA1 was detectable in its cell layer (cytoplasm and ECM) but not in the conditioned medium, indicating the secreted VWA1 was efficiently deposited as ECM. Tubulin and fibronectin (FN) used as loading controls. GuCHL = guanidine hydrochloride extraction. |
|
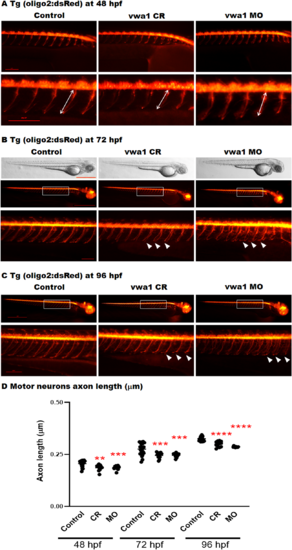
The vwa1 zebrafish model has impaired spinal motor neuron axon development. Transgenic embryos [Tg(olig2:dsRed), expressing red fluorescent oligodendrocytes and motor neurons axons) were injected with vwa1 CRISPR (knockout) or morpholino (MO, knockdown). Both vwa1 models displayed a significantly shorter motor axons in comparison to control axon growth. Representative embryos full-body lateral view and trunk lateral view for the examined groups at: (A) Spinal cord motor neurons examined at 48 hpf [enlarged images below showing the measurement of axon length from the base of the spinal cord to the end of the motor neuron (white arrow)]; (B) 72 hpf, examined spinal motor neurons (white box) and aberrant developing axons (white arrowheads); and (C) 96 hpf. (D) The vwa1 model resulted in impaired motor neurons axon growth in comparison to control axon growth. Spinal motor neurons axon length was measured for 8–10 axons per embryo starting from axon number 5. Vwa1 Crispants (n = 29) and morphants (n = 26) showed significantly shortened axons compared to controls (n = 22). Zebrafish imaging using Lumar 12 microscope (whole body images, scale bar = 40 mm and axon images, scale bar = 10 mm), t-test was used for statistical analysis. Spinal motor neuron images at a magnification of ×150, scale bar = 10 µm. Statistical analysis was performed with GraphPad Prism 8.0. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|