- Title
-
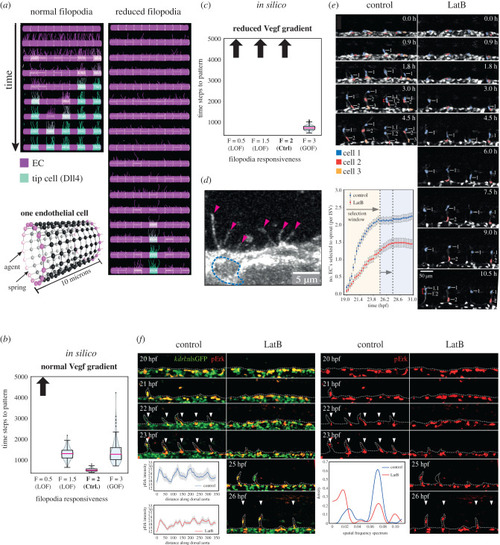
Active perception during angiogenesis: filopodia speed up Notch selection of tip cells in silico and in vivo
- Authors
- Zakirov, B., Charalambous, G., Thuret, R., Aspalter, I.M., Van-Vuuren, K., Mead, T., Harrington, K., Regan, E.R., Herbert, S.P., Bentley, K.
- Source
- Full text @ Phil. Trans. Roy. Soc. Lond., Series B
|
Filopodia loss-of-function (LOF) slows down Notch selection. ( |