- Title
-
Calcium Imaging and the Curse of Negativity
- Authors
- Vanwalleghem, G., Constantin, L., Scott, E.K.
- Source
- Full text @ Front. Neural Circuits
|
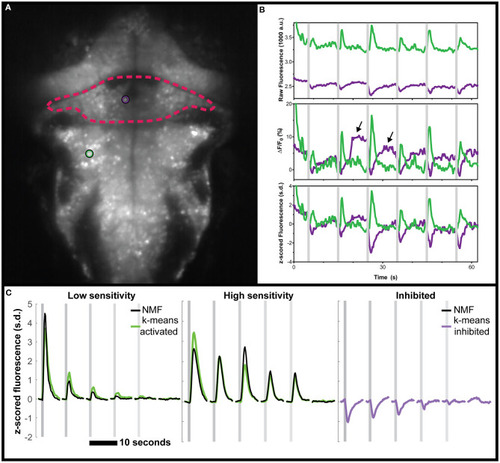
Negative deviations from baseline in real data from the cerebellum of zebrafish, and performance of various analysis tools. |
|
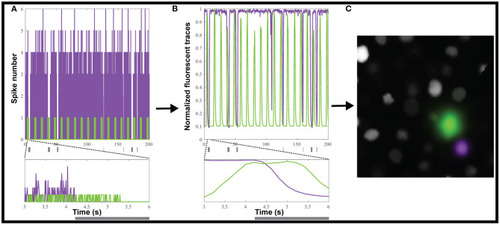
Creating simulated calcium imaging datasets. |
|
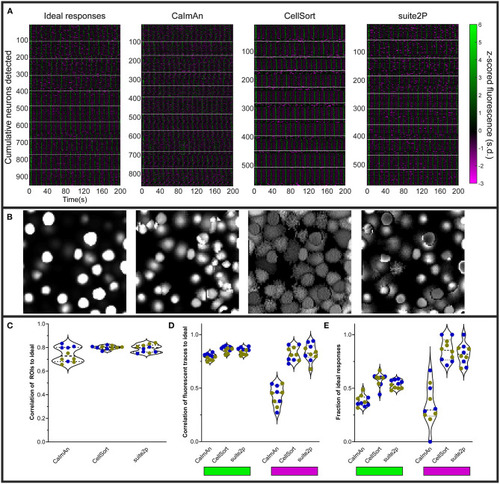
Various analyses' performances on simulated data. |