- Title
-
An improved method for precise genome editing in zebrafish using CRISPR-Cas9 technique
- Authors
- Gasanov, E.V., Jędrychowska, J., Pastor, M., Wiweger, M., Methner, A., Korzh, V.P.
- Source
- Full text @ Mol. Biol. Rep.
|
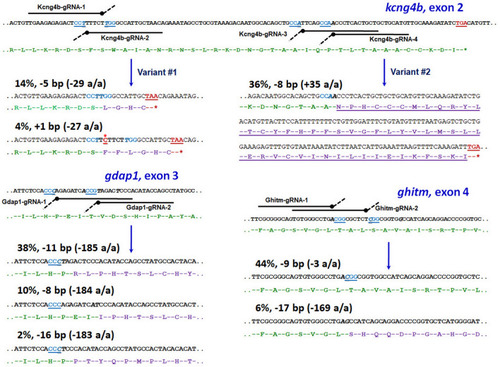
The schematic view of zebrafish ( |
|
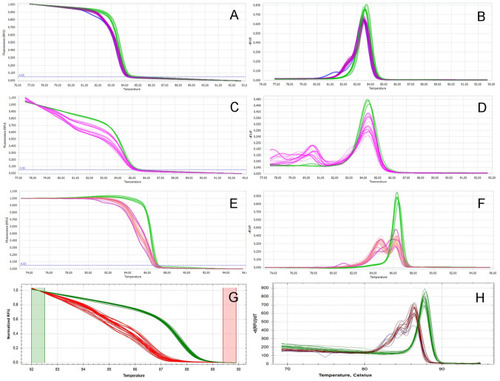
The HRM analysis of the fish generated by precise two gRNAs CRISPR-Cas9 mutagenesis. ( |
|
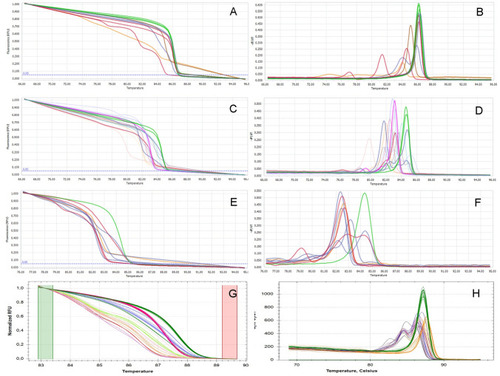
The HRM analysis of the fish generated by one gRNA CRISPR-Cas9 mutagenesis. ( |