- Title
-
MITO-Luc/GFP zebrafish model to assess spatial and temporal evolution of cell proliferation in vivo
- Authors
- de Latouliere, L., Manni, I., Ferrari, L., Pisati, F., Totaro, M.G., Gurtner, A., Marra, E., Pacello, L., Pozzoli, O., Aurisicchio, L., Capogrossi, M.C., Deflorian, G., Piaggio, G.
- Source
- Full text @ Sci. Rep.
|
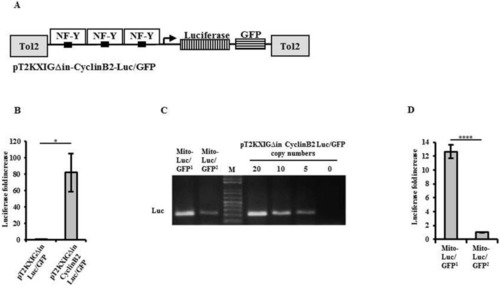
Establishing Tg (cyclin B2:Luc/GFP) MITO-Luc/GFP transgenic zebrafish lines. ( |
|
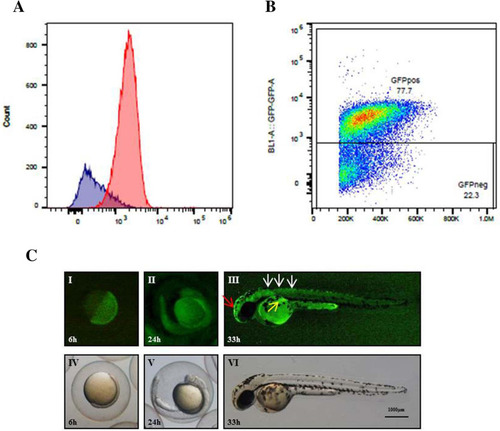
Fluorescence correlates with proliferation in MITO-Luc/GFP zebrafish line. ( |
|
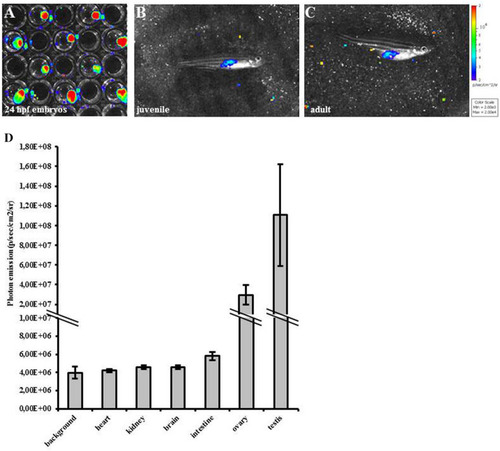
Bioluminescence correlates with proliferation in MITO-Luc/GFP zebrafish line. ( |
|
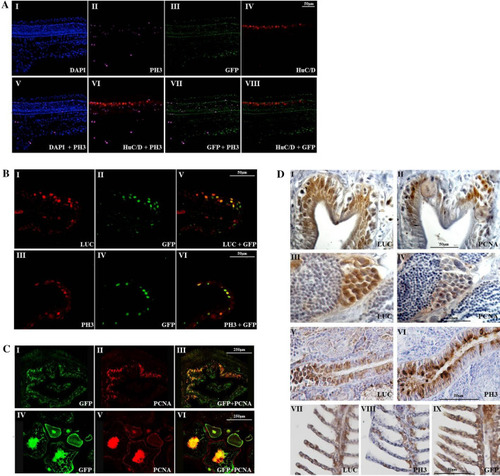
Proliferation markers colocalize with GFP and luciferase proteins in MITO-Luc/GFP zebrafish tissues |
|
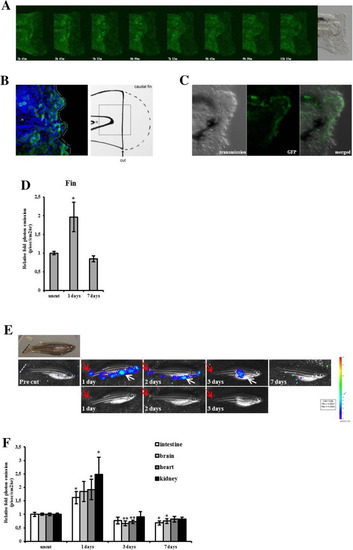
Caudal fin regeneration in MITO-Luc/GFP zebrafish. ( |
|
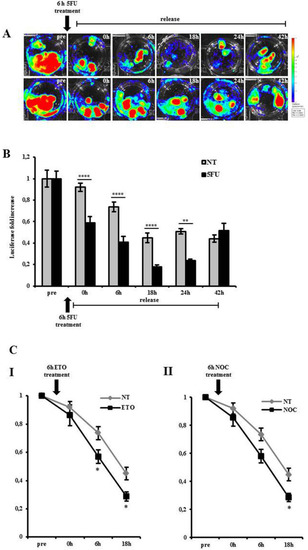
Inhibition of luciferase in zebrafish embryos upon antiproliferative treatments. ( |